Generic model overview¶
The EMOD generic model forms the foundation for all other disease models in EMOD. It provides the fundamental logic for contact-based disease transmission and interventions aimed at controlling the spread of disease that are targeted to individuals or geographic nodes. You can easily add heterogeneity to your simulation by configuring he infectivity of the disease, susceptibility of individuals, and more. The generic model does not contain logic for disease biology, but it can be configured to model a variety of diseases, such as influenza, Ebola, measles, and more. To use the generic model, set the configuration parameter Simulation_Type to GENERIC_SIM.
The figure below demonstrates the main components of the generic EMOD simulation type. Individuals reside in geographic nodes and can migrate from one node to another. Infected individuals shed contagion into a pool that can infect susceptible individuals. You can assign properties to individuals and nodes to vary how transmission or interventions occur.
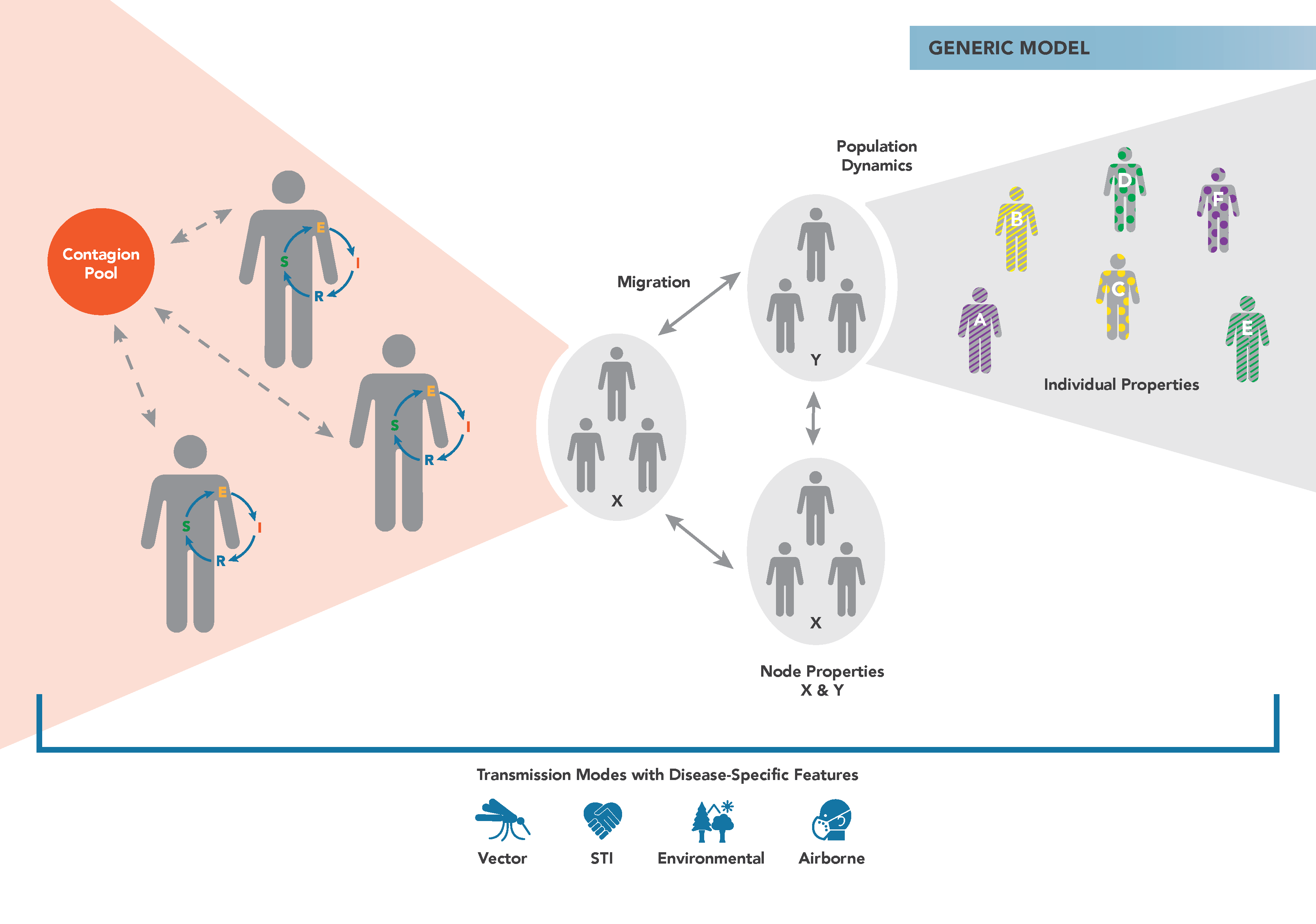
In the generic model, you can add heterogeneous transmission in a population using the Heterogeneous Intra-Node Transmission (HINT) feature. Using this feature, you assign different properties to individuals in a population and differentially scale the infectivity from one group to another. For example, you might have high infectivity between schoolchildren and lower infectivity from the elderly to children. For more information, see Property-based heterogeneous disease transmission (HINT). This feature is not available in most other disease models, where heterogeneity in transmission is handled more mechanistically. For example, the malaria model simulates parasite density, mosquito populations, and biting frequency.
If you are familiar with compartmental models but new to agent-based models, we recommend reviewing the Compartmental models and EMOD section, which compares the mathematics of the differential equations that govern compartmental models with the mathematics of the EMOD agent-based model. It describes how to configure EMOD to model diseases like SI, SIR, or SEIR models. Additionally, review Tutorials and simulation examples for hands-on experience running EMOD simulations.
The configuration of the model regarding infectivity, immune response, and other qualities is handled via several JSON (JavaScript Object Notation) files. For more information, see Overview of EMOD software.
- Compartmental models and EMOD
- Disease outbreaks, reservoirs, and endemicity
- Adding heterogeneity
- Creating campaigns