Models#
IDM builds and uses many different software tools to simulate disease transmission. Selecting the right tool for a given use case will depend on the disease in question and the particular research questions being asked.
Starsim#
The Starsim (or *sim) suite of agent-based models is built in Python and prioritizes usability. Starsim aims to make common tasks easy, while ensuring uncommon tasks are still doable. Starsim leverages the scientific Python ecosystem to keep the code as clear and concise as possible, while still being computationally efficient.
As the Unix wildcard character * implies, this framework can be applied to a variety of different diseases or epidemiological models. All models are open source and we encourage code contributions from those outside of IDM.
Disease/focus |
Code repository |
Model documentation |
Description |
|---|---|---|---|
General disease framework |
Starsim is an agent-based disease modeling framework in which users can design and configure simulations of pathogens that progress over time within each agent and pass from one agent to the next along dynamic transmission networks. The framework explicitly supports co-transmission of multiple pathogens, allowing users to concurrently simulate several diseases while capturing behavioral and biological interactions. |
||
COVID-19 |
Covasim is a stochastic agent-based simulator for performing COVID-19 analyses. These include projections of indicators such as numbers of infections and peak hospital demand. Covasim can also be used to explore the potential impact of different interventions, including social distancing, school closures, testing, contact tracing, quarantine, and vaccination. |
||
Family planning |
FPsim is designed as an open-source tool for family planning research. Its strength stems from a life-course approach, which allows researchers to examine how compounding and temporal effects unfold over women’s lives, and how these individual-level changes lead to macro-level outcomes. |
||
Human papillomavirus (HPV) |
HPVsim is a flexible agent-based model that can be parameterized with country-specific vital dynamics, structured sexual networks, co-transmitting HPV genotypes, B- and T-cell mediated immunity, and high-resolution disease natural history. |
||
STIsim |
STIsim is a stochastic agent-based model, written in Python, for exploring and analyzing sexually transmitted infections. In addition, the repository contains scripts for analyzing model output and tests for testing functionality. |
||
Demographics |
Creates synthetic populations for use with the Starsim models. SynthPops can create generic populations with different network characteristics, as well as synthetic populations that interact in different layers of a multilayer contact network. |
EMOD#
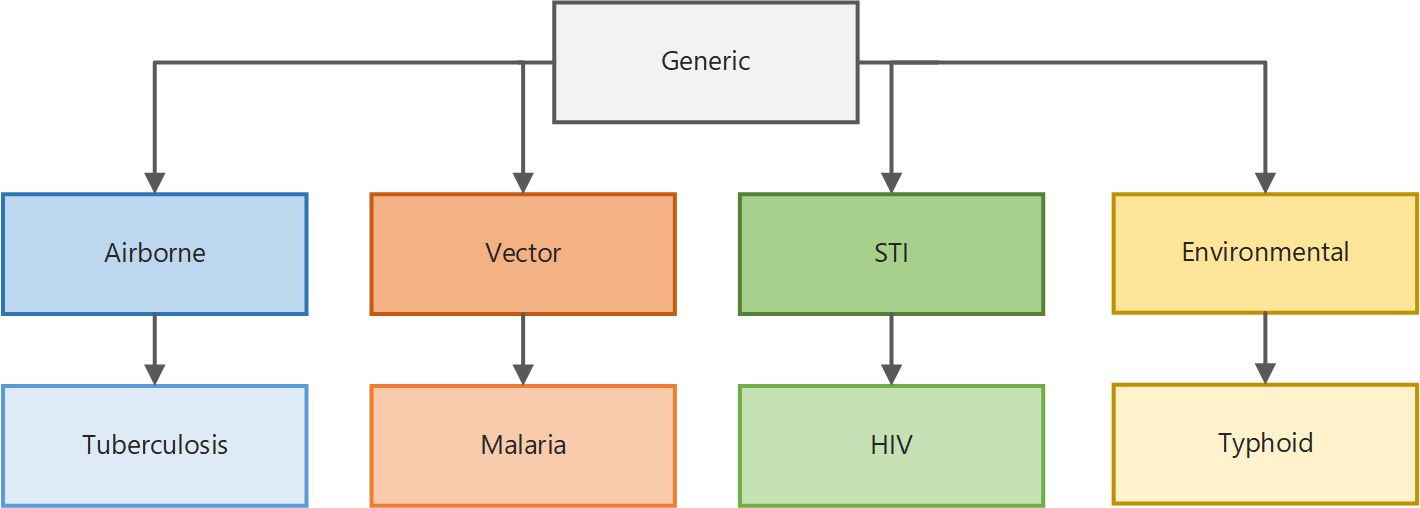
EMOD, an agent-based stochastic model built in C++, is a feature-rich model that utilizes the power of C++ to track the movement, development, health, or other traits of interest of individuals over the course of lifetimes. EMOD uses a layered hierarchy of code features to support simulating different diseases with different modes of transmission.
For example, the generic model (or simulation type) contains base functionality that all other models inherit; the vector model layers on logic for modeling disease transmitted via mosquito; and the malaria model adds functionality specific to the malaria parasite.
Note
Only generic, malaria, and HIV are still being actively developed and supported. Other simulation types and docs are archived for reference.
One key strength of EMOD is the ability to incorporate interventions aimed at controlling or eradicating disease. For example, you can layer multiple interventions, such as bednets, insecticides, and medications, to analyze which combination is most effective at reducing malaria burden.
The latest release is available on GitHub at InstituteforDiseaseModeling/EMOD. The documentation in the table below is for those who want to use EMOD to model disease transmission. For those who want to modify the underlying EMOD C++ code, see the EMOD developer docs.
Disease/focus |
Code repository |
Model documentation |
Description |
|---|---|---|---|
Low complexity disease |
The generic model provides the base model functionality and low-complexity disease transmission, such as measles. |
||
HIV |
The HIV model adds human immunity, within-host viral dynamics, the effects of antiretroviral drugs, and other aspects of HIV virology to simulate HIV disease progression, testing, and treatment. The docs cover EMOD HIV and emodpy-hiv, the Python packaged used to create input files and run simulations. |
||
Malaria |
The malaria model inherits the functionality of the vector model and introduces human immunity, within-host parasite dynamics, effects of antimalarial drugs, and other aspects of malaria biology to simulate malaria transmission. The docs cover EMOD malaria and emodpy-malaria, the Python packaged used to create input files and run simulations. |
||
Tuberculosis (TBHIV) |
The TB model represents the progression of TB disease using a series of state changes as individuals progress from susceptible to latent, to active infection, and then to recovery or death. You can simulate TB transmission with or without including co-infection with HIV. This model is no longer being actively developed or supported. |
||
Typhoid |
The typhoid model adds the progression of typhoid, starting from susceptible then moving through the disease stages of prepatent, subclinical, acute, and chronic, with pathways for recovery and death. The duration of each stage as well as the dosage of contagion exposure can be specified. This model is no longer being actively developed or supported. |
Other models#
Focus |
Code repository |
Model documentation |
Description |
|---|---|---|---|
Stochastic compartmental model |
Compartmental Modeling Software (CMS) is a stochastic compartmental model that is simpler to configure and faster to run than an agent-based model like EMOD, while more suitable to modeling low probability events than a deterministic compartmental model. |
||
Human resources for health |
The PACE-HRH model simulates the healthcare needs of a given population so you can more effectively plan what healthcare worker resources will be necessary to provide a wide range of healthcare services. The model is configured via Excel sheets and run via an R script. |
||
Phylogenetic data |
phyloModels is a Python-based library with utilities for modeling and calibration, with emphasis on exploiting phylogenetic data. |
||
Wastewater and environmental surveillance |
The WES R package provides reproducible functions for collating and analyzing data from wastewater and environmental sampling studies. |
