Malaria infection and immune model¶
To study aspects of malaria infections and population dynamics that do not fit into the framework of a simple SEIRS model with enhancements, EMOD contains a detailed microsolver using within-host parasite dynamics. In the microsolver, the development of clinical and parasitological immunity is tracked through innate and adaptive immune responses to specific antigens. A detailed parasite count over time is tracked for each infected individual. This permits the simulation of measured prevalence over time by slide microscopy, RDTs (Rapid Diagnostic Tests) or other diagnostics. Gametocyte production and decay is included to study the human infectious reservoir.
Immune variables, such as antibody levels for each antigen, inflammatory cytokine levels, and immunological memory are represented by continuous floating-point variables. Most model parameters have a specific physical interpretation and can be rationally constrained by defined experimental measurements.
For a complete list of configuration parameters that are used in the malaria model, see the Configuration parameters.
Within-host parasite dynamics¶
Within the microsolver framework, each new infection begins with a hepatic (liver-stage) latency of fixed duration (7 days) and proceeds through cycles of asexual replication (with a fixed duration of 2 days) and gametocyte production (which takes 10 days). The model accounts for several antigenic components to which the immune system may develop immunity: the merozoite surface protein (MSP) variant, the Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) presented on the surface of the infected red-blood cell (IRBC), and less immunogenic minor surface epitopes (nonspecific epitopes).
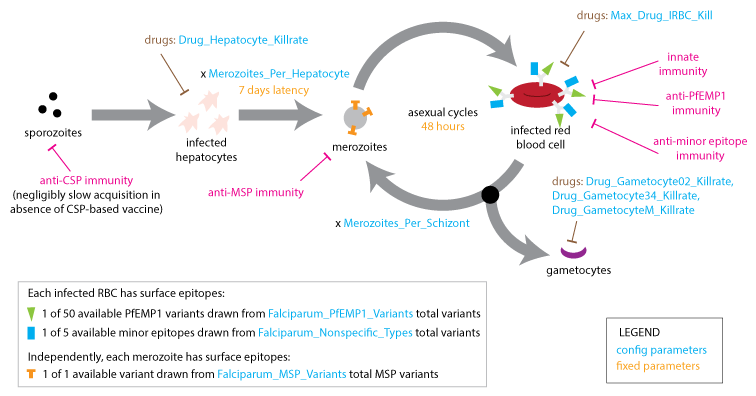
Within-host dynamics¶
Gametocyte development¶
The model advances gametocytes through five stages of development that are characterized by different drug susceptibilities. A fraction of gametocytes die at each developmental stage, and gametocytes can be inactivated by cytokines after uptake in a blood meal. Gametocytes reach maturity after ten days and remain in the bloodstream with a 2.5-day half-life. EMOD does not model acquired immunity to gametocytes.
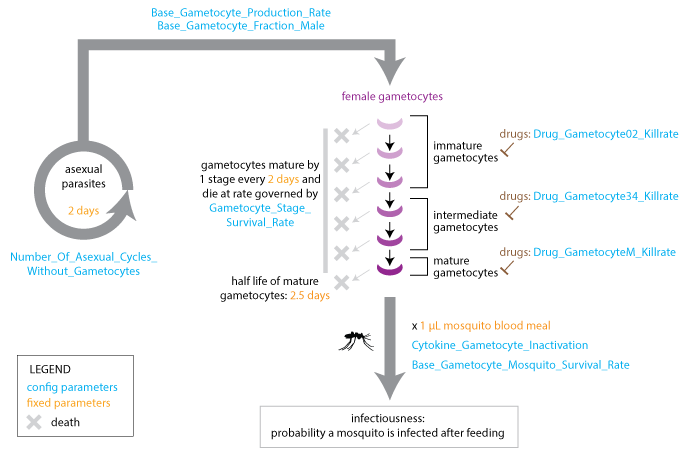
Gametocyte development¶
Modeling a single clonal infection¶
A single clonal infection is modeled with an antigenic repertoire of 50 unique PfEMP-1 variants where each is associated with one of five repeating minor epitopes. In the first asexual cycle, the first five variants are expressed in equal numbers. At each update, the immune system is stimulated by the Infected Red Blood Cell (IRBC) count of each antigenic variant, and concurrently, the IRBC counts are decremented on account of immune and drug killing effects.
At the end of each asexual cycle, the model calculates the fraction of merozoites (16 for each previous IRBC) killed by specific recognition of the MSP variant, and the fraction that is differentiated into male and female gametocytes. To capture the dynamics of the parasite’s immune evasion strategy, the model imposes a constant per-parasite switching rate on the remaining merozoites to advance to subsequent antigenic variants in the repertoire.
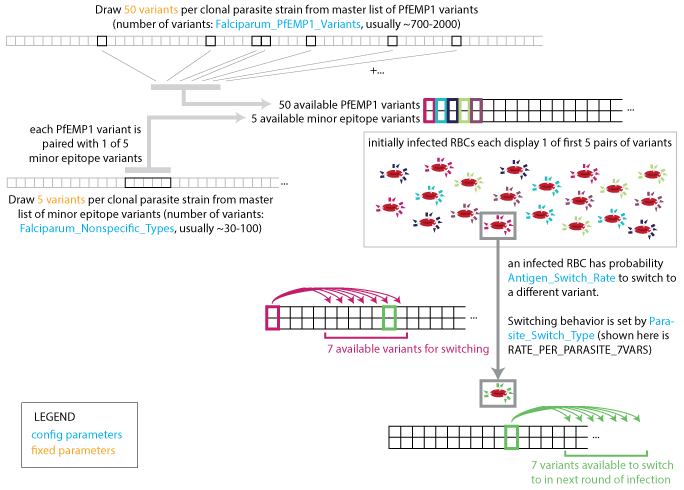
Parasite strain switching¶
Population-level variants¶
The number of population-level variants affect the age-pattern of natural immunity acquisition. The full set of population-level antigenic variants (out of which a single infection’s repertoire is randomly drawn), consists of 100 MSP variants, 20 sets of five minor epitopes, and 1000 PfEMP-1 variants. Some parameters drive the asymptotic levels of adult detected parasitemia while others primarily govern the transition between child and adult detected prevalence rates, and provide that the number of PfEMP-1 variants is substantially more than an individual would experience in a year. A multi-year burn-in period ensures immune initialization dynamics approximate long-term asymptotic behavior where individuals in the simulation build antibody responses to a broad repertoire of parasite antigens appropriate for their age.
Immune response to infection¶
The immune response to infection is characterized by innate inflammatory and specific antibody components that limit maximum parasite density and work to clear infection. As the antibody response increases on a dimensionless scale from 0 to 1, the initial inflammatory response is suppressed. After the antibody response appears, continued antigenic stimulation will drive up the adapted response until that antigenic variant is cleared, at which point both the antibody levels and the capacity to respond will decay over time. The larger the antigenic population, the more infections, and thus time, it takes to acquire broad parasitological immunity.
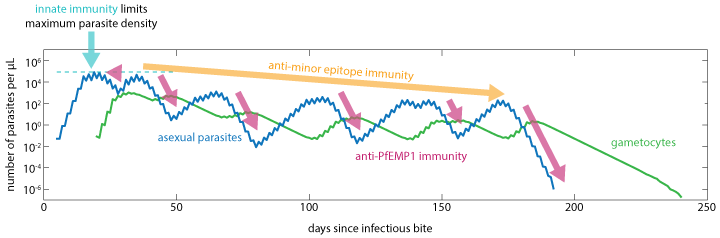
Innate and adaptive immunity work together to limit asexual parasite numbers¶
Innate immune response¶
The innate immune response is modeled to depend on a temporary contribution from a rupturing schizont at the end of each asexual cycle, as well as the concentration of IRBC surface antigens to which an antibody response has not yet been developed. The innate response that is suppressed by the presence of specific antibodies is responsible for driving febrile symptoms and broad-spectrum parasite suppression.
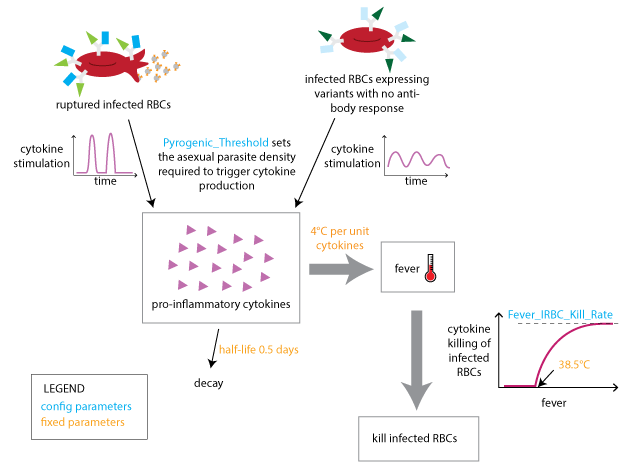
Innate immune response¶
Adaptive immune response¶
The capacity to generate specific antibodies grows in response to the concentration of each novel antigen. Above a capacity threshold level, antibodies are produced in increasing concentration until the corresponding antigenic variant is cleared. At this time, the capacity will decay to a non-zero memory level. The mechanism by which the antibody capacity evolves captures the time delay of specific antibody response on re-infection.
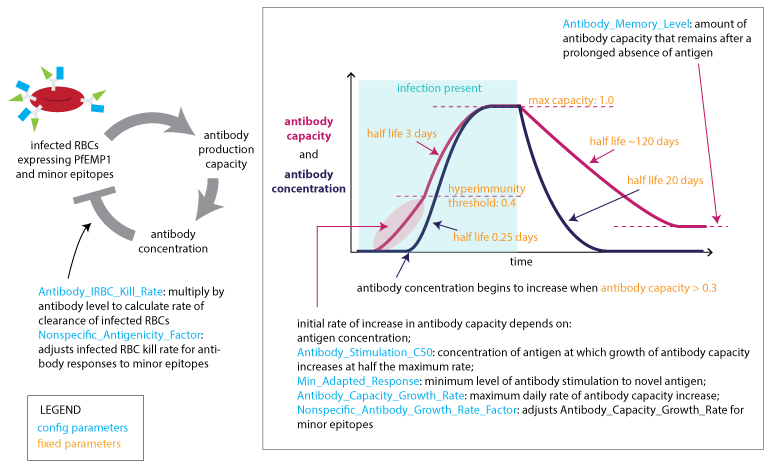
Adaptive immune response to PfEMP1 and minor epitopes¶
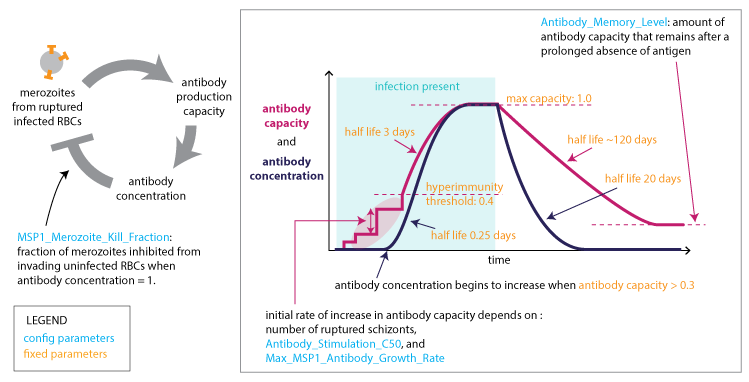
Adaptive immune response to PfEMP1 and minor epitopesAdaptive immune response to MSP antigens¶
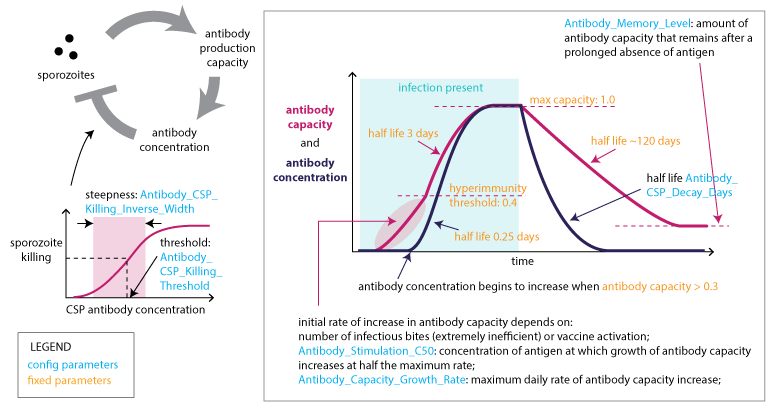
Adaptive immune response to CSP¶
Relevant IDM publications¶
Infection and immunity: Within-host parasite dynamics¶
Lawniczak & Eckhoff, 2016. A computational lens for sexual-stage transmission, reproduction, fitness and kinetics in Plasmodium falciparum. Malaria Journal. 15:487
Cameron, et al., 2015. Defining the relationship between infection prevalence and clinical incidence of Plasmodium falciparum malaria. Nature Communications. 6:8170
Ouedraogo, et al., 2015. Dynamics of the Human Infectious Reservoir for Malaria Determined by Mosquito Feeding Assays and Ultrasensitive Malaria Diagnosis in Burkina Faso. Journal of Infectious Diseases. 213(1):90-99
Gerardin, Ouedraogo, McCarthy, Eckhoff and Wenger, 2015. Characterization of the infectious reservoir of malaria with an agent-based model calibrated to age-stratified parasite densities and infectiousness. Malaria Journal. 14:231
Eckhoff, 2012. Malaria parasite diversity and transmission intensity affect development of parasitological immunity in a mathematical model. Malaria Journal. 11:419
Eckhoff, 2012. P. falciparum Infection Durations and Infectiousness Are Shaped by Antigenic Variation and Innate and Adaptive Host Immunity in a Mathematical Model. PLOS one. 7(9)
Outcrossing of parasite genetics¶
Daniels, et al., 2015. Modeling malaria genomics reveals transmission decline and rebound in Senegal. PNAS. 112(22):7067-7072
Interventions¶
Eckhoff, Wenger, Godfray and Burt, 2016. Impact of mosquito gene drive on malaria elimination in a computational model with explicit spatial and temporal dynamics. PNAS. 114(2)