emod_api.demographics.demographics_base module#
- class emod_api.demographics.demographics_base.DemographicsBase(nodes: List[Node], idref: str, default_node: Node | None = None)[source]#
Bases:
BaseInputFile- Base class for
emod_api:emod_api.demographics.Demographicsand emod_api:emod_api.demographics.DemographicsOverlay.
- DEFAULT_NODE_NAME = 'default_node'#
- exception UnknownNodeException[source]#
Bases:
ValueError
- apply_overlay(overlay_nodes: list)[source]#
- Parameters:
overlay_nodes – Overlay list of nodes over existing nodes in demographics
- Returns:
- send(write_to_this, return_from_forked_sender=False)[source]#
Write data to a file descriptor as specified by the caller. It must be a pipe, a filename, or a file ‘handle’
- Parameters:
write_to_this – File pointer, file path, or file handle.
return_from_forked_sender – Defaults to False. Only applies to pipes. Set to true if caller will handle exiting of fork.
Example:
1) Send over named pipe client code # Named pipe solution 1, uses os.open, not open. import tempfile tmpfile = tempfile.NamedTemporaryFile().name os.mkfifo(tmpfile) fifo_reader = os.open(tmpfile, os.O_RDONLY | os.O_NONBLOCK) fifo_writer = os.open(tmpfile, os.O_WRONLY | os.O_NONBLOCK) demog.send(fifo_writer) os.close(fifo_writer) data = os.read(fifo_reader, int(1e6)) 2) Send over named pipe client code version 2 (forking) import tempfile tmpfile = tempfile.NamedTemporaryFile().name os.mkfifo(tmpfile) process_id = os.fork() # parent stays here, child is the sender if process_id: # reader fifo_reader = open(tmpfile, "r") data = fifo_reader.read() fifo_reader.close() else: # writer demog.send(tmpfile) 3) Send over file. import tempfile tmpfile = tempfile.NamedTemporaryFile().name # We create the file handle and we pass it to the other module which writes to it. with open(tmpfile, "w") as ipc: demog.send(ipc) # Assuming the above worked, we read the file from disk. with open(tmpfile, "r") as ipc: read_data = ipc.read() os.remove(tmpfile)
- Returns:
N/A
- property node_ids#
Return the list of (geographic) node ids.
- property node_count#
Return the number of (geographic) nodes.
- get_node(nodeid: int) Node[source]#
Return the node with node.id equal to nodeid.
- Parameters:
nodeid – an id to use in retrieving the requested Node object. None or 0 for ‘the default node’.
- Returns:
a Node object
- verify_demographics_integrity()[source]#
One stop shopping for making sure a demographics object doesn’t have known invalid settings.
- get_node_by_id(node_id: int) Node[source]#
Returns the Node object requested by its node id.
- Parameters:
node_id – a node_id to use in retrieving the requested Node object. None or 0 for ‘the default node’.
- Returns:
a Node object
- get_nodes_by_id(node_ids: List[int]) Dict[int, Node][source]#
Returns the Node objects requested by their node id.
- Parameters:
node_ids – a list of node ids to use in retrieving Node objects. None or 0 for ‘the default node’.
- Returns:
node entries
- Return type:
a dict with id
- get_node_by_name(node_name: str) Node[source]#
Returns the Node object requested by its node name.
- Parameters:
node_name – a node_name to use in retrieving the requested Node object. None for ‘the default node’.
- Returns:
a Node object
- get_nodes_by_name(node_names: List[str]) Dict[str, Node][source]#
Returns the Node objects requested by their node name.
- Parameters:
node_names – a list of node names to use in retrieving Node objects. None for ‘the default node’.
- Returns:
node entries
- Return type:
a dict with name
- SetMigrationPattern(pattern: str = 'rwd')[source]#
Set migration pattern. Migration is enabled implicitly. It’s unusual for the user to need to set this directly; normally used by emodpy.
- Parameters:
pattern – Possible values are “rwd” for Random Walk Diffusion and “srt” for Single Round Trips.
- SetRoundTripMigration(gravity_factor, probability_of_return=1.0, id_ref='short term commuting migration')[source]#
- Set commuter/seasonal/temporary/round-trip migration rates. You can use the x_Local_Migration configuration
parameter to tune/calibrate.
- Parameters:
gravity_factor – ‘Big G’ in gravity equation. Combines with 1, 1, and -2 as the other exponents.
probability_of_return – Likelihood that an individual who ‘commuter migrates’ will return to the node of origin during the next migration (not timestep). Defaults to 1.0. Aka, travel, shed, return.”
id_ref – Text string that appears in the migration file itself; needs to match corresponding demographics file.
- SetOneWayMigration(rates_path, id_ref='long term migration')[source]#
Set one way migration. You can use the x_Regional_Migration configuration parameter to tune/calibrate.
- Parameters:
rates_path – Path to csv file with node-to-node migration rates. Format is: source (node id),destination (node id),rate.
id_ref – Text string that appears in the migration file itself; needs to match corresponding demographics file.
- SetSimpleVitalDynamics(crude_birth_rate=<emod_api.demographics.DemographicsTemplates.CrudeRate object>, crude_death_rate=<emod_api.demographics.DemographicsTemplates.CrudeRate object>, node_ids=None)[source]#
Set fertility, mortality, and initial age with single birth rate and single mortality rate.
- Parameters:
crude_birth_rate – Birth rate, per year per kiloperson.
crude_death_rate – Mortality rate, per year per kiloperson.
node_ids – Optional list of nodes to limit these settings to.
- SetEquilibriumVitalDynamics(crude_birth_rate=<emod_api.demographics.DemographicsTemplates.CrudeRate object>, node_ids=None)[source]#
Set fertility, mortality, and initial age with single rate and mortality to achieve steady state population.
- Parameters:
crude_birth_rate – Birth rate. And mortality rate.
node_ids – Optional list of nodes to limit these settings to.
- SetEquilibriumVitalDynamicsFromWorldBank(wb_births_df, country, year, node_ids=None)[source]#
Set steady-state fertility, mortality, and initial age with rates from world bank, for given country and year.
- Parameters:
wb_births_df – Pandas dataframe with World Bank birth rate by country and year.
country – Country to pick from World Bank dataset.
year – Year to pick from World Bank dataset.
node_ids – Optional list of nodes to limit these settings to.
- SetDefaultIndividualAttributes()[source]#
NOTE: This is very Measles-ish. We might want to move into MeaslesDemographics
- SetBirthRate(birth_rate, node_ids=None)[source]#
Set Default birth rate to birth_rate. Turn on Vital Dynamics and Births implicitly.
- SetMortalityRate(mortality_rate: CrudeRate, node_ids: List[int] | None = None)[source]#
Set constant mortality rate to mort_rate. Turn on Enable_Natural_Mortality implicitly.
- SetMortalityDistribution(distribution: MortalityDistributionOld | None = None, node_ids: List[int] | None = None)[source]#
Set a default mortality distribution for all nodes or per node. Turn on Enable_Natural_Mortality implicitly.
- Parameters:
distribution – distribution
node_ids – a list of node_ids
- Returns:
None
- SetMortalityDistributionFemale(distribution: MortalityDistributionOld | None = None, node_ids: List[int] | None = None)[source]#
- Set a default female mortality distribution for all nodes or per node. Turn on Enable_Natural_Mortality
implicitly.
- Parameters:
distribution – distribution
node_ids – a list of node_ids
- Returns:
None
- SetMortalityDistributionMale(distribution: MortalityDistributionOld | None = None, node_ids: List[int] | None = None)[source]#
- Set a default male mortality distribution for all nodes or per node. Turn on Enable_Natural_Mortality
implicitly.
- Parameters:
distribution – distribution
node_ids – a list of node_ids
- Returns:
None
- SetMortalityOverTimeFromData(data_csv, base_year, node_ids: List | None = None)[source]#
Set default mortality rates for all nodes or per node. Turn on mortality configs implicitly. You can use the x_Other_Mortality configuration parameter to tune/calibrate.
- Parameters:
data_csv – Path to csv file with the mortality rates by calendar year and age bucket.
base_year – The calendar year the sim is treating as the base.
node_ids – Optional list of node ids to apply this to. Defaults to all.
- Returns:
None
- SetAgeDistribution(distribution: AgeDistributionOld, node_ids: List[int] | None = None)[source]#
Set a default age distribution for all nodes or per node. Sets distribution type to COMPLEX implicitly. :param distribution: age distribution :param node_ids: a list of node_ids
- Returns:
None
- SetDefaultNodeAttributes(birth=True)[source]#
Set the default NodeAttributes (Altitude, Airport, Region, Seaport), optionally including birth, which is most important actually.
- SetDefaultProperties()[source]#
Set a bunch of defaults (age structure, initial susceptibility and initial prevalencec) to sensible values.
- SetDefaultFromTemplate(template, setter_fn=None)[source]#
Add to the default IndividualAttributes using the input template (raw json) and set corresponding config values per the setter_fn. The template should always be constructed by a function in DemographicsTemplates. Eventually this function will be hidden and only accessed via separate application-specific API functions such as the ones below.
- SetEquilibriumAgeDistFromBirthAndMortRates(CrudeBirthRate=<emod_api.demographics.DemographicsTemplates.CrudeRate object>, CrudeMortRate=<emod_api.demographics.DemographicsTemplates.CrudeRate object>, node_ids=None)[source]#
Set the inital ages of the population to a sensible equilibrium profile based on the specified input birth and death rates. Note this does not set the fertility and mortality rates.
- SetInitialAgeExponential(rate=0.0001068, description='')[source]#
Set the initial age of the population to an exponential distribution with a specified rate. :param rate: rate :param description: description, why was this distribution chosen
- SetInitialAgeLikeSubSaharanAfrica(description='')[source]#
Set the initial age of the population to a overly simplified structure that sort of looks like sub-Saharan Africa. This uses the SetInitialAgeExponential. :param description: description, why was this age chosen?
- SetOverdispersion(new_overdispersion_value, nodes: List | None = None)[source]#
Set the overdispersion value for the specified nodes (all if empty).
- SetInitPrevFromUniformDraw(min_init_prev, max_init_prev, description='')[source]#
Set Initial Prevalence (one value per node) drawn from an uniform distribution. :param min_init_prev: minimal initial prevalence :param max_init_prev: maximal initial prevalence :param description: description, why were these parameters chosen?
- AddMortalityByAgeSexAndYear(age_bin_boundaries_in_years: List[float], year_bin_boundaries: List[float], male_mortality_rates: List[List[float]], female_mortality_rates: List[List[float]])[source]#
- SetFertilityOverTimeFromParams(years_region1, years_region2, start_rate, inflection_rate, end_rate, node_ids: List | None = None)[source]#
Set fertility rates that vary over time based on a model with two linear regions. Note that fertility rates use GFR units: babies born per 1000 women of child-bearing age annually. You can use the x_Birth configuration parameter to tune/calibrate.
Refer to the following diagram.
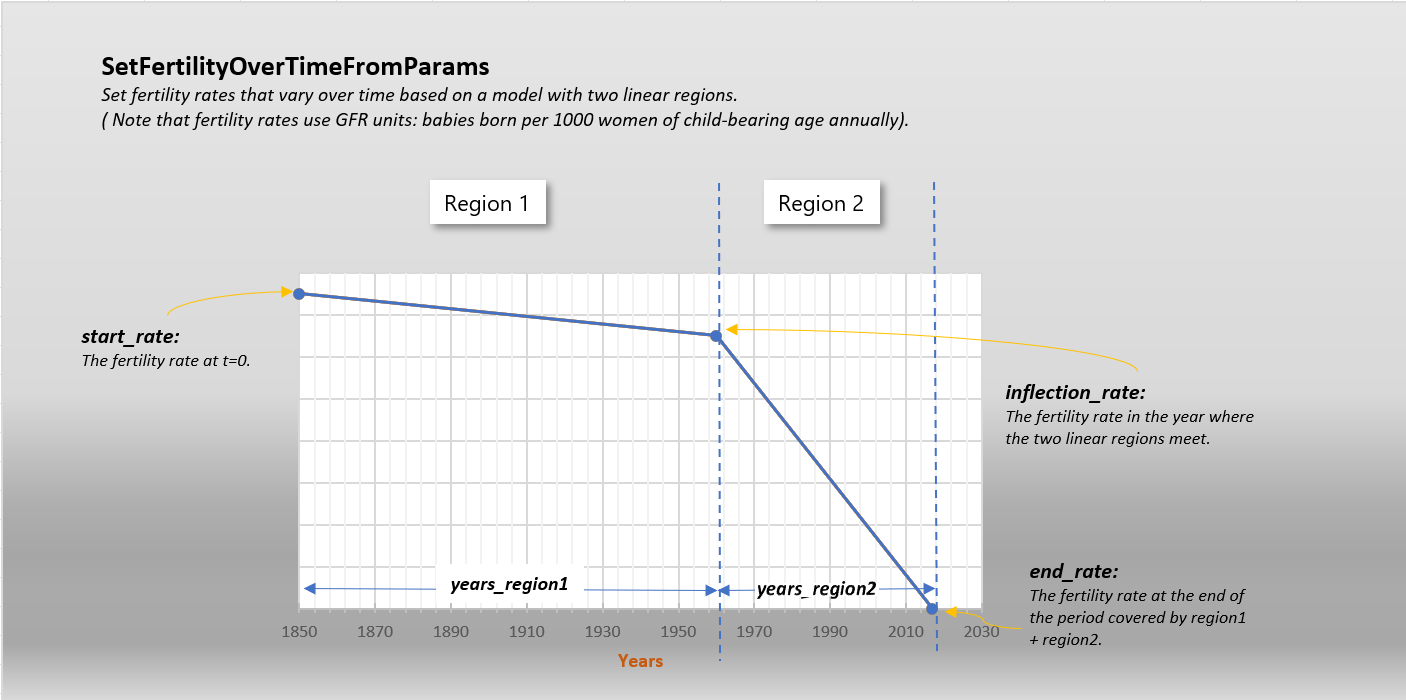
- Parameters:
years_region1 – The number of years covered by the first linear region. So if this represents 1850 to 1960, years_region1 would be 110.
years_region2 – The number of years covered by the second linear region. So if this represents 1960 to 2020, years_region2 would be 60.
start_rate – The fertility rate at t=0.
inflection_rate – The fertility rate in the year where the two linear regions meet.
end_rate – The fertility rate at the end of the period covered by region1 + region2.
node_ids – Optional list of node ids to apply this to. Defaults to all.
- Returns:
rates array (Just in case user wants to do something with them like inspect or plot.)
- infer_natural_mortality(file_male, file_female, interval_fit: List[int | float] | None = None, which_point='mid', predict_horizon=2050, csv_out=False, n=0, results_scale_factor=0.0027397260273972603) [Dict, Dict][source]#
Calculate and set the expected natural mortality by age, sex, and year from data, predicting what it would have been without disease (HIV-only).
- Base class for
