emod_api.demographics.Demographics module#
- emod_api.demographics.Demographics.from_template_node(lat=0, lon=0, pop=1000000, name='Erewhon', forced_id=1)[source]#
Create a single-node
Demographicsinstance from a few parameters.
- emod_api.demographics.Demographics.from_file(base_file)[source]#
Create a
Demographicsinstance from an existing demographics file.
- emod_api.demographics.Demographics.get_node_ids_from_file(demographics_file)[source]#
Get a list of node ids from a demographics file.
- emod_api.demographics.Demographics.get_node_pops_from_params(tot_pop, num_nodes, frac_rural)[source]#
Get a list of node populations from the params used to create a sparsely parameterized multi-node
Demographicsinstance. The first population in the list is the “urban” population and remaning populations are roughly drawn from a log-uniform distribution.- Parameters:
- Returns:
A list containing the urban node population followed by the rural nodes.
- emod_api.demographics.Demographics.from_params(tot_pop=1000000, num_nodes=100, frac_rural=0.3, id_ref='from_params', random_2d_grid=False)[source]#
Create an EMOD-compatible
Demographicsobject with the population and numbe of nodes specified.- Parameters:
tot_pop – The total population.
num_nodes – Number of nodes. Can be defined as a two-dimensional grid of nodes [longitude, latitude]. The distance to the next neighbouring node is 1.
frac_rural – Determines what fraction of the population gets put in the ‘rural’ nodes, which means all nodes besides node 1. Node 1 is the ‘urban’ node.
id_ref – Facility name
random_2d_grid – Create a random distanced grid with num_nodes nodes.
- Returns:
A
Demographicsobject
- emod_api.demographics.Demographics.from_csv(input_file, res=0.008333333333333333, id_ref='from_csv')[source]#
Create an EMOD-compatible
Demographicsinstance from a csv population-by-node file.
- emod_api.demographics.Demographics.from_pop_raster_csv(pop_filename_in, res=0.008333333333333333, id_ref='from_raster', pop_filename_out='spatial_gridded_pop_dir', site='No_Site')[source]#
Take a csv of a population-counts raster and build a grid for use with EMOD simulations. Grid size is specified by grid resolution in arcs or in kilometers. The population counts from the raster csv are then assigned to their nearest grid center and a new intermediate grid file is generated with latitude, longitude and population. This file is then fed to from_csv to generate a demographics object.
- Parameters:
pop_filename_in (str) – The filename of the population-counts raster in CSV format.
res (float, optional) – The grid resolution in arcs or kilometers. Default is 1/120.
id_ref (str, optional) – Identifier reference for the grid. Default is “from_raster”.
pop_filename_out (str, optional) – The output filename for the intermediate grid file. Default is “spatial_gridded_pop_dir”.
site (str, optional) – The site name or identifier. Default is “No_Site”.
- Returns:
py:class`Demographics` object: The generated demographics object based on the grid file.
- Raises:
N/A –
- emod_api.demographics.Demographics.from_pop_csv(pop_filename_in, res=0.008333333333333333, id_ref='from_raster', pop_filename_out='spatial_gridded_pop_dir', site='No_Site')[source]#
Deprecated. Please use from_pop_raster_csv.
- class emod_api.demographics.Demographics.DemographicsBase(nodes: List[Node], idref: str, default_node: Node | None = None)[source]#
Bases:
BaseInputFile- Base class for
emod_api:emod_api.demographics.Demographicsand emod_api:emod_api.demographics.DemographicsOverlay.
- exception UnknownNodeException[source]#
Bases:
ValueError
- apply_overlay(overlay_nodes: list)[source]#
- Parameters:
overlay_nodes – Overlay list of nodes over existing nodes in demographics
- Returns:
- send(write_to_this, return_from_forked_sender=False)[source]#
Write data to a file descriptor as specified by the caller. It must be a pipe, a filename, or a file ‘handle’
- Parameters:
write_to_this – File pointer, file path, or file handle.
return_from_forked_sender – Defaults to False. Only applies to pipes. Set to true if caller will handle exiting of fork.
Example:
1) Send over named pipe client code # Named pipe solution 1, uses os.open, not open. import tempfile tmpfile = tempfile.NamedTemporaryFile().name os.mkfifo(tmpfile) fifo_reader = os.open(tmpfile, os.O_RDONLY | os.O_NONBLOCK) fifo_writer = os.open(tmpfile, os.O_WRONLY | os.O_NONBLOCK) demog.send(fifo_writer) os.close(fifo_writer) data = os.read(fifo_reader, int(1e6)) 2) Send over named pipe client code version 2 (forking) import tempfile tmpfile = tempfile.NamedTemporaryFile().name os.mkfifo(tmpfile) process_id = os.fork() # parent stays here, child is the sender if process_id: # reader fifo_reader = open(tmpfile, "r") data = fifo_reader.read() fifo_reader.close() else: # writer demog.send(tmpfile) 3) Send over file. import tempfile tmpfile = tempfile.NamedTemporaryFile().name # We create the file handle and we pass it to the other module which writes to it. with open(tmpfile, "w") as ipc: demog.send(ipc) # Assuming the above worked, we read the file from disk. with open(tmpfile, "r") as ipc: read_data = ipc.read() os.remove(tmpfile)
- Returns:
N/A
- property node_ids#
Return the list of (geographic) node ids.
- property node_count#
Return the number of (geographic) nodes.
- get_node(nodeid: int) Node[source]#
Return the node with node.id equal to nodeid.
- Parameters:
nodeid – an id to use in retrieving the requested Node object. None or 0 for ‘the default node’.
- Returns:
a Node object
- get_node_by_id(node_id: int) Node[source]#
Returns the Node objects requested by their node id.
- Parameters:
node_id – a node_id to use in retrieving the requested Node object. None or 0 for ‘the default node’.
- Returns:
a Node object
- get_nodes_by_id(node_ids: List[int]) Dict[int, Node][source]#
Returns the Node objects requested by their node id.
- Parameters:
node_ids – a list of node ids to use in retrieving Node objects. None or 0 for ‘the default node’.
- Returns:
node entries
- Return type:
a dict with id
- SetMigrationPattern(pattern: str = 'rwd')[source]#
Set migration pattern. Migration is enabled implicitly. It’s unusual for the user to need to set this directly; normally used by emodpy.
- Parameters:
pattern – Possible values are “rwd” for Random Walk Diffusion and “srt” for Single Round Trips.
- SetRoundTripMigration(gravity_factor, probability_of_return=1.0, id_ref='short term commuting migration')[source]#
- Set commuter/seasonal/temporary/round-trip migration rates. You can use the x_Local_Migration configuration
parameter to tune/calibrate.
- Parameters:
gravity_factor – ‘Big G’ in gravity equation. Combines with 1, 1, and -2 as the other exponents.
probability_of_return – Likelihood that an individual who ‘commuter migrates’ will return to the node of origin during the next migration (not timestep). Defaults to 1.0. Aka, travel, shed, return.”
id_ref – Text string that appears in the migration file itself; needs to match corresponding demographics file.
- SetOneWayMigration(rates_path, id_ref='long term migration')[source]#
Set one way migration. You can use the x_Regional_Migration configuration parameter to tune/calibrate.
- Parameters:
rates_path – Path to csv file with node-to-node migration rates. Format is: source (node id),destination (node id),rate.
id_ref – Text string that appears in the migration file itself; needs to match corresponding demographics file.
- SetSimpleVitalDynamics(crude_birth_rate=<emod_api.demographics.DemographicsTemplates.CrudeRate object>, crude_death_rate=<emod_api.demographics.DemographicsTemplates.CrudeRate object>, node_ids=None)[source]#
Set fertility, mortality, and initial age with single birth rate and single mortality rate.
- Parameters:
crude_birth_rate – Birth rate, per year per kiloperson.
crude_death_rate – Mortality rate, per year per kiloperson.
node_ids – Optional list of nodes to limit these settings to.
- SetEquilibriumVitalDynamics(crude_birth_rate=<emod_api.demographics.DemographicsTemplates.CrudeRate object>, node_ids=None)[source]#
Set fertility, mortality, and initial age with single rate and mortality to achieve steady state population.
- Parameters:
crude_birth_rate – Birth rate. And mortality rate.
node_ids – Optional list of nodes to limit these settings to.
- SetEquilibriumVitalDynamicsFromWorldBank(wb_births_df, country, year, node_ids=None)[source]#
Set steady-state fertility, mortality, and initial age with rates from world bank, for given country and year.
- Parameters:
wb_births_df – Pandas dataframe with World Bank birth rate by country and year.
country – Country to pick from World Bank dataset.
year – Year to pick from World Bank dataset.
node_ids – Optional list of nodes to limit these settings to.
- SetIndividualAttributesWithFertMort(crude_birth_rate=<emod_api.demographics.DemographicsTemplates.CrudeRate object>, crude_mort_rate=<emod_api.demographics.DemographicsTemplates.CrudeRate object>)[source]#
- AddIndividualPropertyAndHINT(Property: str, Values: List[str], InitialDistribution: List[float] | None = None, TransmissionMatrix: List[List[float]] | None = None, Transitions: List | None = None, node_ids: List[int] | None = None, overwrite_existing: bool = False) None[source]#
Add Individual Properties, including an optional HINT configuration matrix.
Individual properties act as ‘labels’ on model agents that can be used for identifying and targeting subpopulations in campaign elements and reports. E.g. model agents may be given a property (‘Accessibility’) that labels them as either having access to health care (value: ‘Yes’) or not (value: ‘No’).
Property-based heterogeneous disease transmission (HINT) is available for generic, environmental, typhoid, airborne, or TBHIV simulations as other simulation types have parameters for modeling the heterogeneity of transmission. By default, transmission is assumed to occur homogeneously among the population within a node.
- Note: EMOD requires individual property key and values (Property and Values args) to be the same across all
nodes. The individual distributions of individual properties (InitialDistribution) can vary acros nodes.
- Documentation of individual properties and HINT:
https://docs.idmod.org/projects/emod-generic/en/latest/model-properties.html https://docs.idmod.org/projects/emod-generic/en/latest/model-hint.html
- Parameters:
Property – a new individual property key to add (if property already exists an exception is raised unless overwrite_existing is True).
Values – the valid values of the new property key
InitialDistribution – The fractional initial distribution of each valid Values entry. Order must match Values argument.
TransmissionMatrix – HINT transmission matrix.
node_ids – The node ids to apply changes to. None or 0 means the ‘Defaults’ node.
overwrite_existing – Determines if an error is thrown if the IP is found pre-existing at a specified node. False: throw exception. True: overwrite the existing property.
- Returns:
None
- AddAgeDependentTransmission(Age_Bin_Edges_In_Years: List | None = None, TransmissionMatrix: List[List[float]] | None = None)[source]#
Set up age-based HINT. Since ages are a first class property of an agent, Age_Bin is a special case of HINT. We don’t specify a distribution, but we do specify the age bin edges, in units of years. So if Age_Bin_Edges_In_Years = [0, 10, 65, -1] it means you’ll have 3 age buckets: 0-10, 10-65, & 65+. Always ‘book-end’ with 0 and -1.
- Parameters:
Age_Bin_Edges_In_Years – array (or list) of floating point values, representing the age bucket bounderies.
TransmissionMatrix – 2-D array of floating point values, representing epi connectedness of the age buckets.
- SetDefaultIndividualAttributes()[source]#
NOTE: This is very Measles-ish. We might want to move into MeaslesDemographics
- SetBirthRate(birth_rate, node_ids=None)[source]#
Set Default birth rate to birth_rate. Turn on Vital Dynamics and Births implicitly.
- SetMortalityRate(mortality_rate: CrudeRate, node_ids: List[int] | None = None)[source]#
Set constant mortality rate to mort_rate. Turn on Enable_Natural_Mortality implicitly.
- SetMortalityDistribution(distribution: MortalityDistribution | None = None, node_ids: List[int] | None = None)[source]#
Set a default mortality distribution for all nodes or per node. Turn on Enable_Natural_Mortality implicitly.
- Parameters:
distribution – distribution
node_ids – a list of node_ids
- Returns:
None
- SetVitalDynamicsFromWHOFile(pop_dat_file: Path, base_year: int, start_year: int = 1950, max_daily_mort: float = 0.01, mortality_rate_x_values: list = [0.6, 1829.5, 1829.6, 3659.5, 3659.6, 5489.5, 5489.6, 7289.5, 7289.6, 9119.5, 9119.6, 10949.5, 10949.6, 12779.5, 12779.6, 14609.5, 14609.6, 16439.5, 16439.6, 18239.5, 18239.6, 20069.5, 20069.6, 21899.5, 21899.6, 23729.5, 23729.6, 25559.5, 25559.6, 27389.5, 27389.6, 29189.5, 29189.6, 31019.5, 31019.6, 32849.5, 32849.6, 34679.5, 34679.6, 36509.5, 36509.6, 38339.5], years_per_age_bin: int = 5)[source]#
Build demographics from UN World Population data. :param pop_dat_file: path to UN World Population data file :param base_year: Base year/Reference year :param start_year: Read in the pop_dat_file starting with year ‘start_year’ :param years_per_age_bin: The number of years in one age bin, i.e. in one row of the UN World Population data file :param max_daily_mort: Maximum daily mortality rate :param mortality_rate_x_values: The distribution of non-disease mortality for a population.
- Returns:
IndividualAttributes, NodeAttributes
- SetMortalityDistributionFemale(distribution: MortalityDistribution | None = None, node_ids: List[int] | None = None)[source]#
- Set a default female mortality distribution for all nodes or per node. Turn on Enable_Natural_Mortality
implicitly.
- Parameters:
distribution – distribution
node_ids – a list of node_ids
- Returns:
None
- SetMortalityDistributionMale(distribution: MortalityDistribution | None = None, node_ids: List[int] | None = None)[source]#
- Set a default male mortality distribution for all nodes or per node. Turn on Enable_Natural_Mortality
implicitly.
- Parameters:
distribution – distribution
node_ids – a list of node_ids
- Returns:
None
- SetMortalityOverTimeFromData(data_csv, base_year, node_ids: List | None = None)[source]#
Set default mortality rates for all nodes or per node. Turn on mortality configs implicitly. You can use the x_Other_Mortality configuration parameter to tune/calibrate.
- Parameters:
data_csv – Path to csv file with the mortality rates by calendar year and age bucket.
base_year – The calendar year the sim is treating as the base.
node_ids – Optional list of node ids to apply this to. Defaults to all.
- Returns:
None
- SetAgeDistribution(distribution: AgeDistribution, node_ids: List[int] | None = None)[source]#
Set a default age distribution for all nodes or per node. Sets distribution type to COMPLEX implicitly. :param distribution: age distribution :param node_ids: a list of node_ids
- Returns:
None
- SetDefaultNodeAttributes(birth=True)[source]#
Set the default NodeAttributes (Altitude, Airport, Region, Seaport), optionally including birth, which is most important actually.
- SetDefaultProperties()[source]#
Set a bunch of defaults (age structure, initial susceptibility and initial prevalencec) to sensible values.
- SetDefaultPropertiesFertMort(crude_birth_rate=<emod_api.demographics.DemographicsTemplates.CrudeRate object>, crude_mort_rate=<emod_api.demographics.DemographicsTemplates.CrudeRate object>)[source]#
Set a bunch of defaults (birth rates, death rates, age structure, initial susceptibility and initial prevalence) to sensible values.
- SetDefaultFromTemplate(template, setter_fn=None)[source]#
Add to the default IndividualAttributes using the input template (raw json) and set corresponding config values per the setter_fn. The template should always be constructed by a function in DemographicsTemplates. Eventually this function will be hidden and only accessed via separate application-specific API functions such as the ones below.
- SetNodeDefaultFromTemplate(template, setter_fn)[source]#
Add to the default NodeAttributes using the input template (raw json) and set corresponding config values per the setter_fn. The template should always be constructed by a function in DemographicsTemplates. Eventually this function will be hidden and only accessed via separate application-specific API functions such as the ones below.
- SetEquilibriumAgeDistFromBirthAndMortRates(CrudeBirthRate=<emod_api.demographics.DemographicsTemplates.CrudeRate object>, CrudeMortRate=<emod_api.demographics.DemographicsTemplates.CrudeRate object>, node_ids=None)[source]#
Set the inital ages of the population to a sensible equilibrium profile based on the specified input birth and death rates. Note this does not set the fertility and mortality rates.
- SetInitialAgeExponential(rate=0.0001068, description='')[source]#
Set the initial age of the population to an exponential distribution with a specified rate. :param rate: rate :param description: description, why was this distribution chosen
- SetInitialAgeLikeSubSaharanAfrica(description='')[source]#
Set the initial age of the population to a overly simplified structure that sort of looks like sub-Saharan Africa. This uses the SetInitialAgeExponential. :param description: description, why was this age chosen?
- SetOverdispersion(new_overdispersion_value, nodes: List | None = None)[source]#
Set the overdispersion value for the specified nodes (all if empty).
- SetConstantSusceptibility()[source]#
Set the initial susceptibilty for each new individual to a constant value of 1.0.
- SetInitPrevFromUniformDraw(min_init_prev, max_init_prev, description='')[source]#
Set Initial Prevalence (one value per node) drawn from an uniform distribution. :param min_init_prev: minimal initial prevalence :param max_init_prev: maximal initial prevalence :param description: description, why were these parameters chosen?
- SetConstantRisk(risk=1, description='')[source]#
Set the initial risk for each new individual to the same value, defaults to full risk :param risk: risk :param description: description, why was this parameter chosen?
- SetHeteroRiskUniformDist(min_risk=0, max_risk=1)[source]#
Set the initial risk for each new individual to a value drawn from a uniform distribution.
- SetHeteroRiskLognormalDist(mean=1.0, sigma=0)[source]#
Set the initial risk for each new individual to a value drawn from a log-normal distribution.
- SetHeteroRiskExponDist(mean=1.0)[source]#
Set the initial risk for each new individual to a value drawn from an exponential distribution.
- AddMortalityByAgeSexAndYear(age_bin_boundaries_in_years: List[float], year_bin_boundaries: List[float], male_mortality_rates: List[List[float]], female_mortality_rates: List[List[float]])[source]#
- SetFertilityOverTimeFromParams(years_region1, years_region2, start_rate, inflection_rate, end_rate, node_ids: List | None = None)[source]#
Set fertility rates that vary over time based on a model with two linear regions. Note that fertility rates use GFR units: babies born per 1000 women of child-bearing age annually. You can use the x_Birth configuration parameter to tune/calibrate.
Refer to the following diagram.
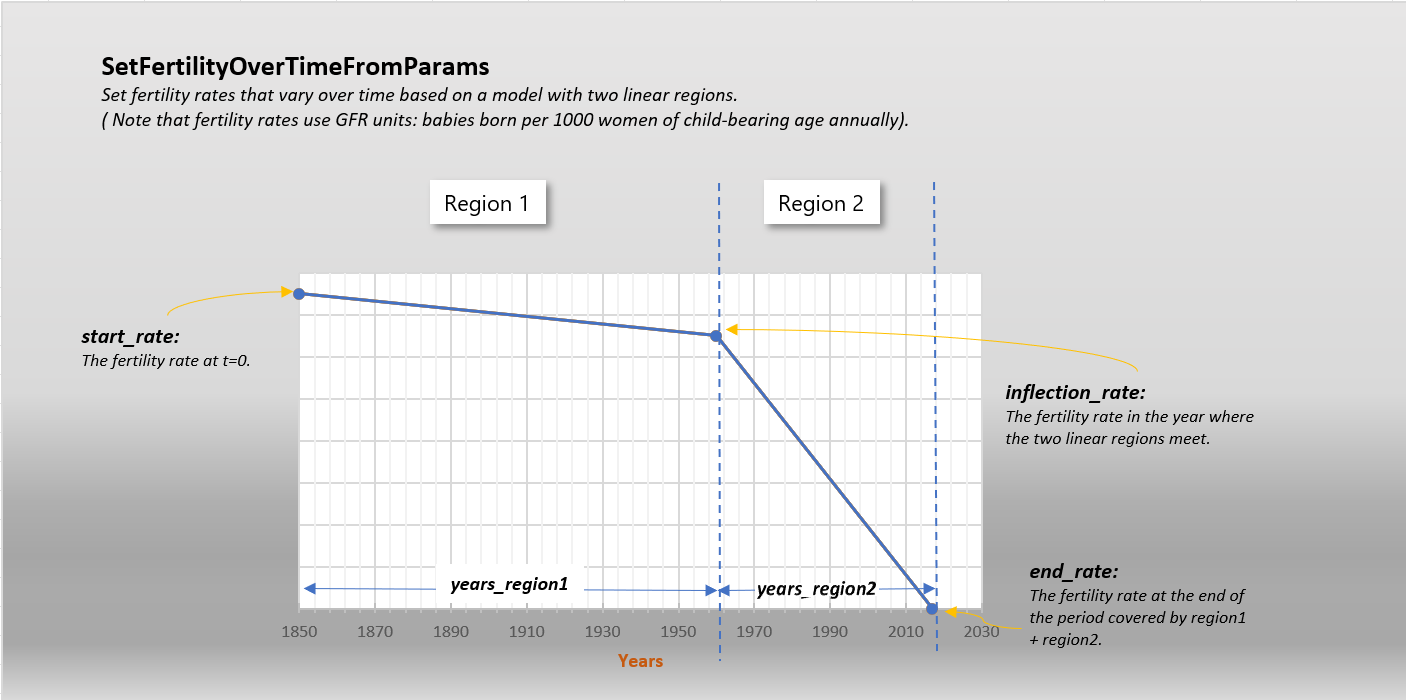
- Parameters:
years_region1 – The number of years covered by the first linear region. So if this represents 1850 to 1960, years_region1 would be 110.
years_region2 – The number of years covered by the second linear region. So if this represents 1960 to 2020, years_region2 would be 60.
start_rate – The fertility rate at t=0.
inflection_rate – The fertility rate in the year where the two linear regions meet.
end_rate – The fertility rate at the end of the period covered by region1 + region2.
node_ids – Optional list of node ids to apply this to. Defaults to all.
- Returns:
rates array (Just in case user wants to do something with them like inspect or plot.)
- infer_natural_mortality(file_male, file_female, interval_fit: List[int | float] | None = None, which_point='mid', predict_horizon=2050, csv_out=False, n=0, results_scale_factor=0.0027397260273972603) [Dict, Dict][source]#
Calculate and set the expected natural mortality by age, sex, and year from data, predicting what it would have been without disease (HIV-only).
- Base class for
- class emod_api.demographics.Demographics.DemographicsOverlay(nodes: list | None = None, idref: str | None = None, individual_attributes=None, node_attributes=None)[source]#
Bases:
DemographicsBaseIn contrast to class
emod_api:emod_api.demographics.Demographicsthis class does not set any defaults. It inherits fromemod_api:emod_api.demographics.DemographicsBaseso all functions that can be used to create demographics can also be used to create an overlay file. Parameters can be changed/set specifically by passing node_id, individual attributes, and individual attributes to the constructor.
- class emod_api.demographics.Demographics.Demographics(nodes: List[Node], idref: str = 'Gridded world grump2.5arcmin', base_file: str | None = None, default_node: Node | None = None)[source]#
Bases:
DemographicsBaseThis class is a container of data necessary to produce a EMOD-valid demographics input file. It can be initialized from an existing valid demographics.joson type file or from an array of valid Nodes.
