Software overview#
The Institute for Disease Modeling (IDM) develops detailed simulations of disease transmission through the use of quantitative software modeling. The agent-based model, Epidemiological MODeling software (EMOD), helps determine the combination of health policies and intervention strategies that can lead to disease eradication. EMOD calculates how diseases may spread in particular areas and is used to analyze the effects of current and future health policies and intervention strategies. It supports infectious disease campaign planning, data gathering, new product development, and policy decisions. IDM shares this modeling software with the research community to advance the understanding of disease dynamics.
EMOD is quite different from a deterministic compartmental model, which uses an ordinary differential equation (ODE) to determine the rates at which proportions of the population move from each compartment (or disease state) to another. EMOD is an agent-based model (ABM) that simulates the simultaneous interactions of agents in an effort to recreate complex phenomena. Each agent, whether it is a human or a vector, can be assigned a variety of properties to represent age, gender, parasite density, and much more. Their behavior and interactions with one another are determined by using decision rules based on their properties. These models have strong predictive power and are able to leverage spatial and temporal dynamics.
EMOD is also stochastic, meaning that there is randomness built into the model. Infection and recovery processes are represented as probabilistic Bernoulli random draws. In other words, when a susceptible person comes into contact with a pathogen, they are not guaranteed to become infected. Instead, you can imagine flipping a coin that has a λ chance of coming up tails S(t) times and every person who gets a head is considered infected. This randomness better approximates what happens in reality. It also means that you must run many simulations to determine the probability of particular outcomes.
Simulation types#
EMOD currently supports the following simulation types for modeling a variety of diseases:
Generic disease (GENERIC_SIM), which can be used for modeling a variety of diseases such as influenza or measles
Vector-borne diseases (VECTOR_SIM), which can be used for modeling vector-borne diseases such as dengue
Malaria (MALARIA_SIM), which adds features specific to malaria biology and treatment
Sexually transmitted infections (STI_SIM), which adds features for sexual relationship networks
HIV (HIV_SIM), which adds features specific to HIV biology and treatment
The illustration below shows how the simulation types are built upon one another. With a few exceptions, all parameters available to configure the simulation in the generic simulation type are inherited by the vector simulation type. The vector simulation type adds additional parameters specific to the biology of vector-borne diseases, which in turn are inherited by the malaria simulation type, which adds parameters specific to malaria biology and treatment, and so on. Therefore, depending on the simulation type you select, different parameters are available for you to use. In addition, simulation types for broader classes of disease can be extended to build your own disease-specific model.
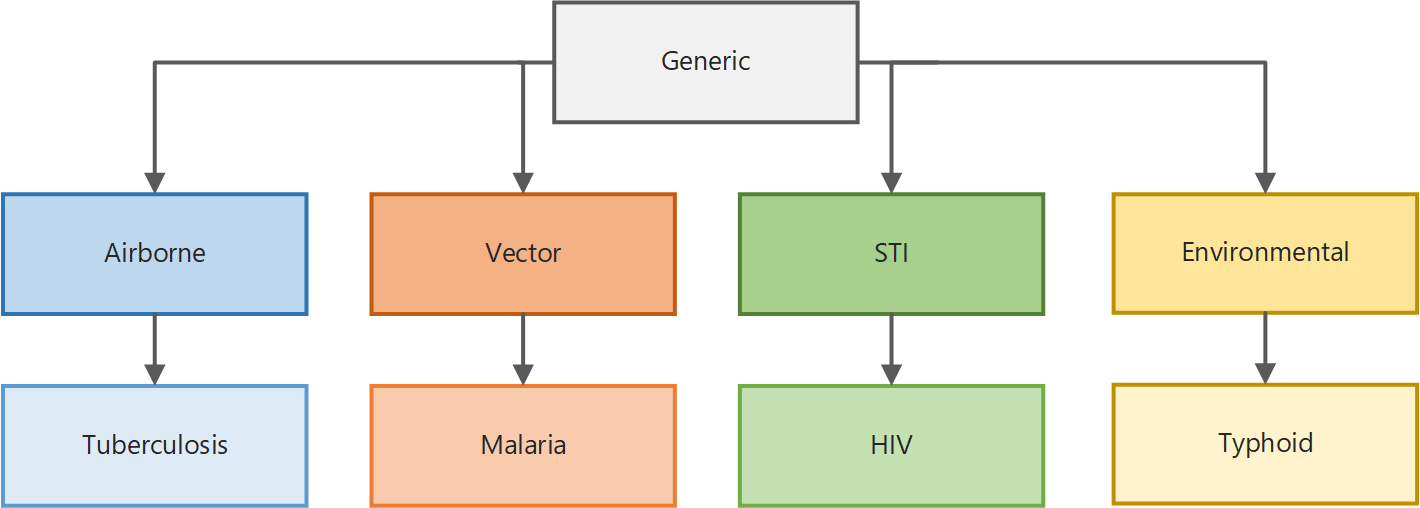
Simulation type inheritance#
EMOD utilizes object-oriented programming design principles such as interfaces, factories, and observers to achieve a highly modular architecture, thus enabling comparisons of structural assumptions and different levels of model detail. For example, parameters specific to the intrahost biology of disease progression and to behavioral propensities that drive health care seeking have been separated into different inheritance classes. Thus, modules or sub-modules can be interchanged while leaving other portions of the model intact.
EMOD can produce statistically significant results over a broad set of parameters and scenarios. One strength of EMOD is the ability to incorporate interventions aimed at controlling or eradicating disease. Quantitative analysis of the simulated output enables disease eradication efforts to be more driven by data. The IDM research team has published many articles related to modeling and the modeling concepts underpinning EMOD. For a list of published articles, see IDM Publications.
This section provides an overview of EMOD and files needed to run simulations. The architecture diagram below shows, at a high level, how the system functions. If you run simulations in parallel on a multi-node cluster, there is also a Message Passing Interface (MPI) component used to pass data between multiple instances of EMOD.
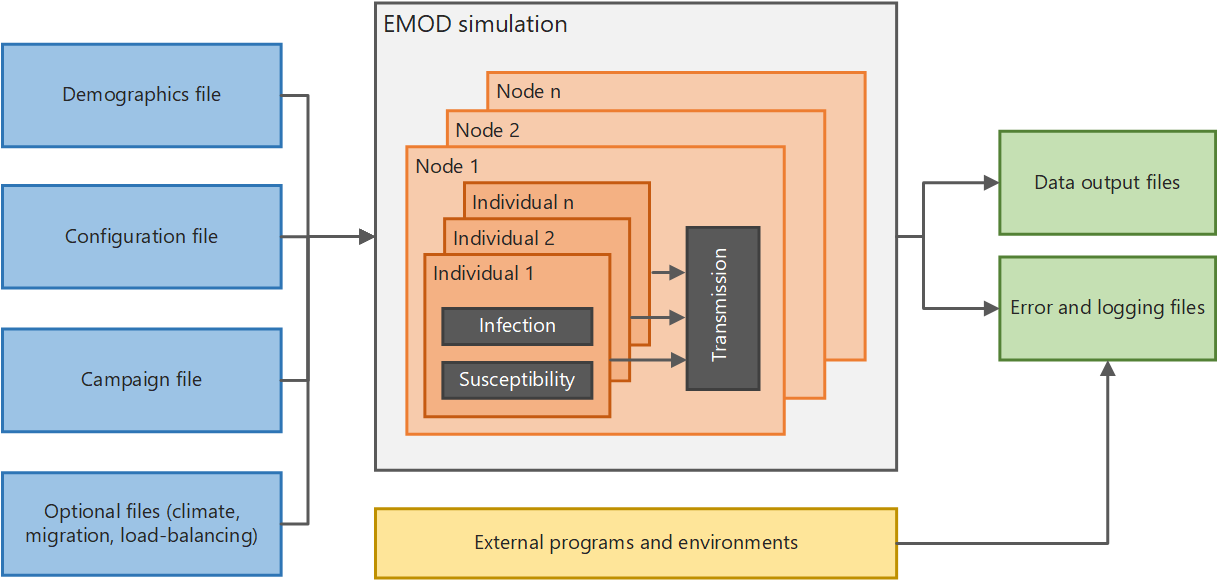
High-level EMOD system architecture#
- Input files
- Running a simulation
- Output files (reports)
- Custom reporters
- Error and logging files
- Binned output report
- Demographic summary output report
- Inset chart output report
- Property output report
- Event counter report (ReportEventCounter.json)
- Health events and interventions report
- Human migration tracking report (ReportHumanMigrationTracking.csv)
- Node demographics report (ReportNodeDemographics.csv)
- Spatial output report
