T4 - Diseases#
An interactive version of this notebook is available on Google Colab or Binder.
Diseases are the cornerstone of almost any Starsim analysis. This is where you add details about what disease you are modeling, including transmissibility, natural history, and mortality outcomes.
NOTE 1: The version of Starsim was originally conceived as a model for co-circulating STIs, and the current version of the codebase has much more developed models of STIs (including syphilis and HIV) than it does for other pathogens. However, the library of available diseases will keep expanding with time, and we will update these tutorials as it does.
NOTE 2: Starsim descended from a family of similar agent-based models that share common architecture and design principles: Covasim, HPVsim, and FPsim. If you are especially interested in modeling COVID-19, HPV and cervical cancer, or family planning, you may be better off using one of these pre-existing models. For all other diseases/health conditions, Starsim is probably your best bet.
Modifying a disease#
Much like sims or networks, a Disease can be customized by passing in a pars dictionary containing parameters. We’ve already seen examples of this, but
[1]:
import starsim as ss
sir = ss.SIR(dur_inf=10, beta=0.2, init_prev=0.4, p_death=0.2)
sim = ss.Sim(n_agents=2_000, diseases=sir, networks='random')
sim.run()
Initializing sim with 2000 agents
Running 2000.0 ( 0/51) (0.00 s) ———————————————————— 2%
Running 2010.0 (10/51) (0.05 s) ••••———————————————— 22%
Running 2020.0 (20/51) (0.08 s) ••••••••———————————— 41%
Running 2030.0 (30/51) (0.11 s) ••••••••••••———————— 61%
Running 2040.0 (40/51) (0.15 s) ••••••••••••••••———— 80%
Running 2050.0 (50/51) (0.18 s) •••••••••••••••••••• 100%
[1]:
Sim(n=2000; 2000—2050; networks=randomnet; diseases=sir)
We already saw that this model creates results that are stored in sim.results.sir. The results can also be directly accessed via sir.results.
For more detail on any of the diseases that are in the Starsim library of diseases, please refer to the docstrings and source code of the disease files.
For more detail on making your own disease, please refer to the developer tutorial.
Adding multiple diseases#
You can add multiple diseases to the same simulation, like so. Here we are making use of a “connector”. A connector is a module in Starsim that tells you how two things relate to one another - in this case, how HIV modifies a person’s transmissibility and susceptibility to gonorrhea and vice versa. Unlike dieases, networks, interventions, etc., connectors don’t have any pre-specified location in the sim. Instead, they can be placed wherever they make the most sense (for example, a connector that mediated how two networks behaved might be placed at the beginning or end of the list of networks; for diseases, it might be placed at the beginning or end of the list of diseases).
[2]:
import sciris as sc
import starsim as ss
class simple_hiv_ng(ss.Module):
""" Simple connector whereby rel_sus to NG doubles if CD4 count is <200"""
def __init__(self, pars=None, label='HIV-Gonorrhea', **kwargs):
super().__init__()
self.define_pars(
rel_trans_hiv = 2,
rel_trans_aids = 5,
rel_sus_hiv = 2,
rel_sus_aids = 5,
)
self.update_pars(pars, **kwargs)
return
def step(self):
""" Specify how HIV increases NG rel_sus and rel_trans """
people = self.sim.people
people.gonorrhea.rel_sus[people.hiv.cd4 < 500] = self.pars.rel_sus_hiv
people.gonorrhea.rel_sus[people.hiv.cd4 < 200] = self.pars.rel_sus_aids
people.gonorrhea.rel_trans[people.hiv.cd4 < 500] = self.pars.rel_trans_hiv
people.gonorrhea.rel_trans[people.hiv.cd4 < 200] = self.pars.rel_trans_aids
return
# Make HIV
hiv = ss.HIV(
beta = {'mf': [0.0008, 0.0004]}, # Specify transmissibility over the MF network
init_prev = 0.05,
)
# Make gonorrhea
ng = ss.Gonorrhea(
beta = {'mf': [0.05, 0.025]}, # Specify transmissibility over the MF network
init_prev = 0.025,
)
# Make the sim, including a connector betweeh HIV and gonorrhea:
n_agents = 5_000
sim = ss.Sim(n_agents=n_agents, networks='mf', diseases=[simple_hiv_ng(), hiv, ng])
sim.run()
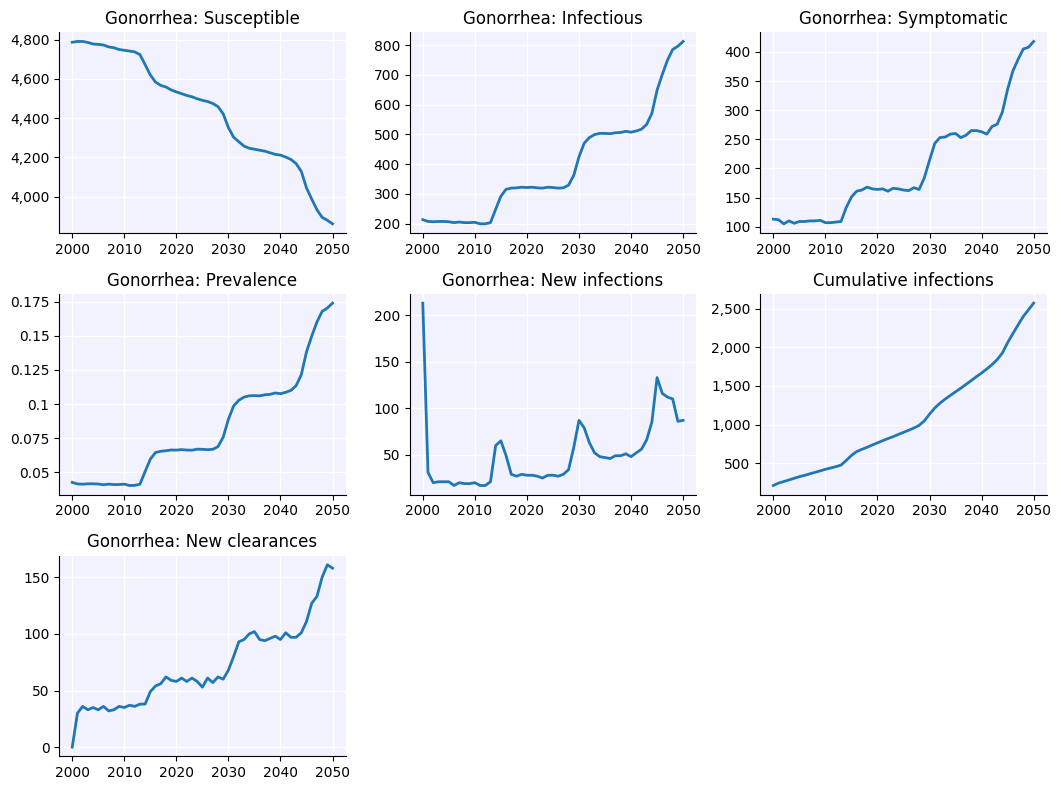
sim.plot('hiv')
sim.plot('gonorrhea')
Initializing sim with 5000 agents
Running 2000.0 ( 0/51) (0.00 s) ———————————————————— 2%
Running 2010.0 (10/51) (0.09 s) ••••———————————————— 22%
Running 2020.0 (20/51) (0.19 s) ••••••••———————————— 41%
Running 2030.0 (30/51) (0.28 s) ••••••••••••———————— 61%
Running 2040.0 (40/51) (0.37 s) ••••••••••••••••———— 80%
Running 2050.0 (50/51) (0.46 s) •••••••••••••••••••• 100%
Figure(1066.67x800)
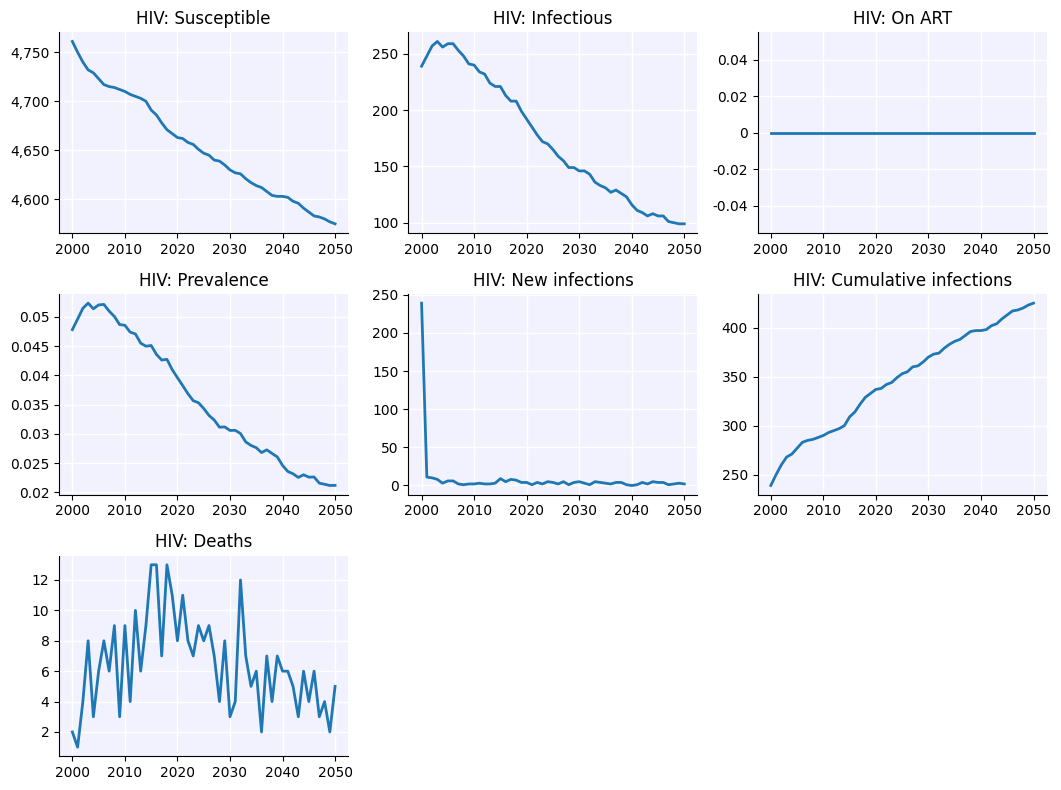
Figure(1066.67x800)