Simulation core components#
The simulation component contains core functionality that models the behavior of a disease without any interventions and extended functionality to include migration, climate, or other input data to create a more realistic simulation. Disease transmission may be more or less complex depending on the disease being modeled.
Warning
If you modify the source code to add or remove configuration or campaign parameters, you may need to update the code used to produce the schema. You must also verify that your simulations are still scientifically valid.
Each generic EMOD simulation contains the following core base classes:
- Simulation
Created by the simulation controller via a SimulationFactory with each run of EMOD.
- Node
Corresponds to a geographic area. Each simulation maintains a collection of one or more nodes.
- IndividualHuman
Represents a human being. Creates Susceptibility and Infection objects for the collection of individuals it maintains. The file name that defines this class is “Individual” and you may see it likewise shortened in diagrams.
- Susceptibility
Manages an individual’s immunity.
- Infection
Represents an individual’s infection with a disease.
For generic simulations, each node has a homogeneous contagion pool. Every individual in the node sheds disease into the pool and acquires disease from the pool. You can add heterogeneous disease transmission within a node by enabling and configuring the Heterogeneous Intra-Node Transmission (HINT) feature. For more information, see Property-based heterogeneous disease transmission (HINT).
The relationship between these classes is captured in the following figure.
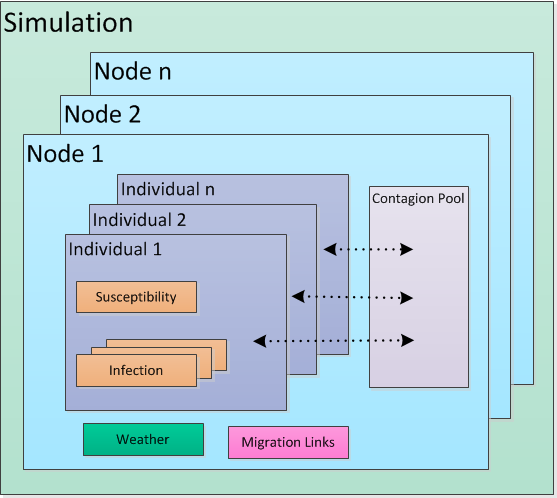
Simulation components#
After the simulation is initialized, all objects in the simulation are updated at each time step, typically a single day. Each object implements a method Update that advances the state of the objects it contains, as follows:
Controller updates Simulation
Simulation updates Nodes
Node updates IndividualHuman
IndividualHuman updates Susceptibility, Infections, and InterventionsContainer
InterventionsContainer updates Interventions
Simulation#
As a stochastic model, EMOD uses a random number seed for all simulations. The Simulation object has a data member (RNG) that is an object maintaining the state of the random number generator for the parent Simulation object. The only generator currently supported is pseudo-DES. The random seed is initialized from the configuration parameter Run_Number and from the process MPI rank. All child objects needing access to the RNG must be provided an appropriate (context) pointer by their owners.
The Simulation class contains the following methods:
Method |
Description |
|---|---|
Populate() |
Initializes the simulation. The Populate method initializes the simulation using both the configuration file and the demographic files. Populate calls through to populateFromDemographics to enable the Simulation object to create one or many Node objects populated with IndividualHumans as dictated by the demographics file, in conjunction with the sampling mode and value dictated by the configuration file. If the configuration file indicates that a migration and a climate model are to be used, those input file are also read. Populate also initializes all Reporters. |
Update() |
Advances the state of nodes. |
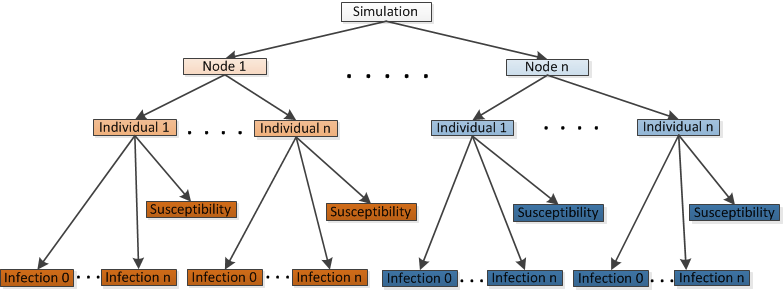
Simulation object hierarchy#
For multi-core parallelization, the demographics file is read in order on each process and identity of each node and is compared with a policy assigning nodes to processes embodied in objects implementing InitialLoadBalancingScheme. If the initial load balancing scheme allows a node for the current rank, the node is created via addNewNodeFromDemographics. After all nodes have been created and propagated, the NodeRankMaps are merged across all processes. See Load-balancing files for more information.
Node#
Nodes are model abstractions that represent a population of individuals that interact in a way that does not depend on their geographic location. However, they represent a geographic location with latitude and longitude coordinates, climate information, migration links to other nodes, and miscellaneous demographic information. The Node is always the container for IndividualHumans and the contagion pool. The Node provides important capabilities for how IndividualHumans are created and managed. It can also contain a Climate object and Migration links if those features are enabled. The climate and migration settings are initialized based on the information in the input data files.
The Node class contains the following methods:
Method |
Description |
|---|---|
PopulateFromDemographics() |
The entry point that invokes populateNewIndividualsFromDemographics(InitPop), which adds individuals to a simulation and initializes them. PopulateNewIndividualsFromBirth() operates similarly, but can use different distributions for demographics and initial infections. |
Update() |
Advances the state of individuals. |
updateInfectivity() |
The workhorse of the simulation, it processes the list of all individuals attached to the Node object and updates the force of infection data members in the contagion pool object. It calls a base class function updatePopulationStatistics, which processes all individuals, sets the counters for prevalence reporting, and calls IndividualHuman::GetInfectiousness for all IndividualHuman objects. The code in GetInfectiousness governs the interaction of the IndividualHuman with the contagion pool object. The rest of the code in updateInfectivity processes the contagion beyond individual contributions. This can include decay of persisting contagion, vector population dynamics, seasonality, etc. This is also where the population-summed infectivity must be scaled by the population in the case of density-independent transmission. |
updateVitalDynamics() |
Manages community level vital dynamics, primarily births, since deaths occur at the individual level. |
By default, an IndividualHuman object is created, tracked, and updated for every person within a node. To reduce memory usage and processing time, you may want to sample such that each IndividualHuman object represents multiple people. There are several different sampling strategies implemented, with different strategies better suited for different simulations. See Sampling for more information.
If migration is enabled, at the end of the Node update, the Node moves all migrating individuals to a separate migration queue for processing. Once the full simulation time step is completed, all migrating individuals are moved from the migration queue and added to their destination nodes.
IndividualHuman#
The IndividualHuman class corresponds to human beings within the simulation. Individuals are always contained by a Node object. Each IndividualHuman object may represent one or more human beings, depending on the sampling strategy and value chosen.
The IndividualHuman class contains components for susceptibility, infection, and interventions. Infection and Susceptibility cooperate to represent the detailed dynamics of infection and immune mechanisms. Every IndividualHuman contains a Susceptibility object that represents the state of the immune system over time. Only an infected IndividualHuman contains an Infection object, and may contain multiple Infection objects.. Susceptibility is passed to initialize the infection immunology in Infection::InitInfectionImmunology(). The state of an individual’s susceptibility and infection are updated with Update() methods. Disease-specific models have additional derived classes with properties and methods to represent specifics of the disease biology.
The InterventionsContainer is the mediating structure for how interventions interrupt disease transmission or progression. Campaign distribution results in an Intervention object being added to an individual’s InterventionsContainer, where it remains unless and until it is removed. When an IndividualHuman calls Update(), the InterventionsContainer is updated and its effects are applied to the IndividualHuman. These effects are used in the individual, infection, and susceptibility update operations. If migration is enabled, at the end of each individual’s update step EMOD checks if the individual is scheduled for migration (IndividualHuman::CheckForMigration()), setting a flag accordingly.
The IndividualHuman class contains the following methods:
Method |
Description |
|---|---|
Update() |
Advances the state of both the infection and the immune system and then registers any necessary changes in an individual’s state resulting from those dynamics (that is, death, paralysis, or clearance). It also updates intrinsic vital dynamics, intervention effects, migration, and exposure to infectivity of the appropriate social network. |
ExposeToInfectivity() |
Passes the IndividualHuman to the ExposeIndividual() function if it is exposed to infectivity at a time step. |
UpdateInfectiousness() |
Advances the quantity of contagion deposited to the contagion pool by an IndividualHuman at each time step of their infectious period. This is explained in more detail below. |
Disease transmission#
Transmission of disease is mediated through a pool mechanism which tracks abstract quantities of contagion. The pool mediates individual acquisition and transmission of infections as well as external processes that modify the infectivity dynamics external to individuals. The pool provides basic mechanisms for depositing, decaying, and querying quantities of contagion which are associated with a specific StrainIdentity. The pool internally manages a separate ContagionPopulation for each possible antigen identity. ContagionPopulations have further structure and manage an array of contagion quantities for each substrain identity.
Each IndividualHuman has a sampling weight \(W_i\) and a total infectiousness \(X_i\), the rate at which contacts with the infectious individual become infected. This rate can be modified by transmission-reducing immunity or heterogeneous contact rates, which are gathered in \(Y_{i,transmit}\), and the transmission-reducing effects of interventions, such as transmission- blocking vaccines in the factor \(Z_{i,transmit}\). The sampling weight \(W_i\) is not included in the probability of acquiring a new infection. Sample particles are simulated as single individuals, their weighting \(W_i\) is used to determine their effects upon the rest of the simulation.The total infectiousness \(T\) of the local human population is then calculated as:
For simulation of population density-independent transmission, individual infectiousness \(X_i\) includes the average contact rate of the infectious individual, so this total infectiousness is divided by the population \(P\) to get the force of infection \(FI=\frac{T}{P}\) for each individual in the population. The base rate of acquisition of new infections per person is then \(FI\), which can be modified for each individual \(I\) by their characteristics \(Y_{i,acquire}\) and interventions \(Z_{i,acquire}\). Over a time step \(\Delta t\), the probability of an individual acquiring a new infection is then:
A new infection receives an incubation period and infectious period from the input configuration (a constant at present, but possibly from specified distributions in the future) and a counter tracks the full latency, which is possible when simulating individual particles. After the incubation period, the counter for the infectious period begins, during which time the infection contributes to the individual’s infectiousness \(X_i\).
