Developer tutorial: Analyzers#
An interactive version of this notebook is available on Google Colab or Binder.
Reporting results#
Each Starsim module can have its own results, which get added to the full list of results in the Sim object. For example, the ss.Pregnancy module adds results like sim.results.pregnancy.pregnant, and the ss.HIV module adds results like sim.results.hiv.new_infections. If you are writing your own module, you can add whatever custom results you want. However, another option is to create an Analyzer to store results that you might need for one particular analysis but won’t need
all the time. An Analyzer is very similar to other Starsim modules in its structure, but the general idea of an analyzer is that it gets called at the end of a timestep, and reports of the state of things after everything else has been updated without changing any of the module states itself.
Simple usage#
For simple reporting, it’s possible to use a single function as an analyzer. In this case, the function receives a single argument, sim, which it has full access to. For example, if you wanted to know the number of connections in the network on each timestep, you could write a small analyzer as follows:
[1]:
import starsim as ss
import matplotlib.pyplot as plt
# Store the number of edges
n_edges = []
def count_edges(sim):
""" Print out the number of edges in the network on each timestep """
network = sim.networks[0] # Get the first network
n = len(network)
n_edges.append(n)
print(f'Number of edges for network {network.name} on step {sim.ti}: {n}')
return
# Create the sim
pars = dict(
diseases='sis',
networks = 'mf',
analyzers = count_edges,
demographics = True,
)
# Run the sim
sim = ss.Sim(pars).run()
sim.plot()
# Plot the number of edges
plt.figure()
plt.plot(sim.timevec, n_edges)
plt.title('Number of edges over time')
plt.show()
Initializing sim with 10000 agents
Running 2000.0 ( 0/51) (0.00 s) ———————————————————— 2%
Number of edges for network mfnet on step 0: 3756
Number of edges for network mfnet on step 1: 3721
Number of edges for network mfnet on step 2: 3688
Number of edges for network mfnet on step 3: 3663
Number of edges for network mfnet on step 4: 3630
Number of edges for network mfnet on step 5: 3594
Number of edges for network mfnet on step 6: 3551
Number of edges for network mfnet on step 7: 3524
Number of edges for network mfnet on step 8: 3503
Number of edges for network mfnet on step 9: 3462
Running 2010.0 (10/51) (0.63 s) ••••———————————————— 22%
Number of edges for network mfnet on step 10: 3425
Number of edges for network mfnet on step 11: 3400
Number of edges for network mfnet on step 12: 3366
Number of edges for network mfnet on step 13: 3326
Number of edges for network mfnet on step 14: 3302
Number of edges for network mfnet on step 15: 3289
Number of edges for network mfnet on step 16: 3272
Number of edges for network mfnet on step 17: 3274
Number of edges for network mfnet on step 18: 3271
Number of edges for network mfnet on step 19: 3271
Running 2020.0 (20/51) (0.70 s) ••••••••———————————— 41%
Number of edges for network mfnet on step 20: 3260
Number of edges for network mfnet on step 21: 3249
Number of edges for network mfnet on step 22: 3233
Number of edges for network mfnet on step 23: 3246
Number of edges for network mfnet on step 24: 3236
Number of edges for network mfnet on step 25: 3226
Number of edges for network mfnet on step 26: 3217
Number of edges for network mfnet on step 27: 3202
Number of edges for network mfnet on step 28: 3180
Number of edges for network mfnet on step 29: 3188
Running 2030.0 (30/51) (0.76 s) ••••••••••••———————— 61%
Number of edges for network mfnet on step 30: 3198
Number of edges for network mfnet on step 31: 3202
Number of edges for network mfnet on step 32: 3195
Number of edges for network mfnet on step 33: 3194
Number of edges for network mfnet on step 34: 3193
Number of edges for network mfnet on step 35: 3194
Number of edges for network mfnet on step 36: 3185
Number of edges for network mfnet on step 37: 3197
Number of edges for network mfnet on step 38: 3196
Number of edges for network mfnet on step 39: 3191
Running 2040.0 (40/51) (0.83 s) ••••••••••••••••———— 80%
Number of edges for network mfnet on step 40: 3195
Number of edges for network mfnet on step 41: 3201
Number of edges for network mfnet on step 42: 3202
Number of edges for network mfnet on step 43: 3179
Number of edges for network mfnet on step 44: 3167
Number of edges for network mfnet on step 45: 3161
Number of edges for network mfnet on step 46: 3157
Number of edges for network mfnet on step 47: 3157
Number of edges for network mfnet on step 48: 3147
Number of edges for network mfnet on step 49: 3116
Running 2050.0 (50/51) (0.89 s) •••••••••••••••••••• 100%
Number of edges for network mfnet on step 50: 3093
Figure(933.333x700)
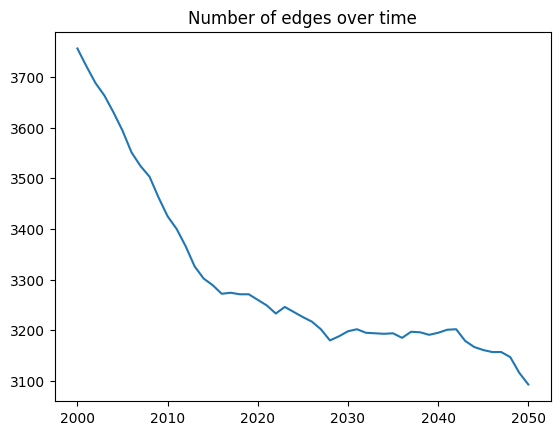
Is that what you expected it to look like? The reason it looks like that is that initially, agents die (either from aging or from disease), reducing the number of edges. New agents are being born, but they don’t participate in male-female networks until the age of debut – which is 15 years old by default, which is why the trend reverses (and tracks population size) after 2015. This illustrates the importance of model burn-in!
Advanced usage#
Suppose we wanted to create an analyzer that would report on the number of new HIV infections in pregnant women:
[2]:
import starsim as ss
import pandas as pd
class HIV_preg(ss.Analyzer):
def __init__(self, *args, **kwargs):
super().__init__(*args, **kwargs)
return
def init_results(self):
super().init_results()
self.define_results(
ss.Result('new_infections_pregnancy'),
)
return
def step(self):
sim = self.sim
ti = sim.ti
hiv = sim.diseases.hiv
pregnant = sim.demographics.pregnancy.pregnant
newly_infected = hiv.ti_infected == ti
self.results['new_infections_pregnancy'][ti] = len((newly_infected & pregnant).uids)
return
pregnancy = ss.Pregnancy(pars=dict(fertility_rate=pd.read_csv('test_data/nigeria_asfr.csv')))
hiv = ss.HIV(beta={'mfnet':[0.5,0.25]})
sim = ss.Sim(diseases=hiv, networks='mfnet', demographics=pregnancy, analyzers=HIV_preg())
sim.run()
print(f'Total infections among pregnant women: {sim.results.hiv_preg.new_infections_pregnancy.sum()}')
Initializing sim with 10000 agents
Running 2000.0 ( 0/51) (0.00 s) ———————————————————— 2%
Running 2010.0 (10/51) (0.10 s) ••••———————————————— 22%
Running 2020.0 (20/51) (0.23 s) ••••••••———————————— 41%
Running 2030.0 (30/51) (0.39 s) ••••••••••••———————— 61%
Running 2040.0 (40/51) (0.56 s) ••••••••••••••••———— 80%
Running 2050.0 (50/51) (0.74 s) •••••••••••••••••••• 100%
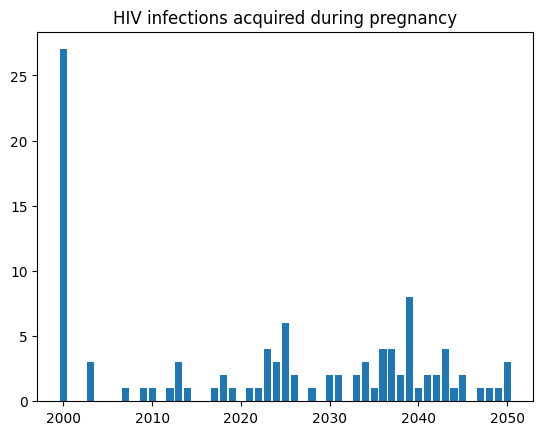
Total infections among pregnant women: 106.0
Analyzers are ideal for adding custom results, and because they get added to the sim in the same way as any other result, they also get automatically exported in the same format, e.g. using sim.to_df().
Here’s a plot of the results from our HIV in pregnancy analyzer:
[3]:
import matplotlib.pyplot as plt
res = sim.results.hiv_preg
plt.figure()
plt.bar(res.timevec, res.new_infections_pregnancy)
plt.title('HIV infections acquired during pregnancy')
plt.show()