T1 - Getting started#
Starsim is a highly flexible framework for creating agent-based models. Starsim was designed by a team with expertise in infectious diseases, so many of the examples that we’ll cover throughout these tutorials are examples of infectious diseases. However, you can also use Starsim for modeling other things, like non-communicable diseases (NCDs) or family planning and maternal health.
Installing and getting started with Starsim is quite simple.
To install, just type pip install starsim. If it worked, you should be able to import Starsim with import starsim as ss.
The basic design philosophy of Starsim is: common tasks should be simple. Since Starsim is most commonly used for modeling infectious diseases, everything related to that should be straightforward. The most common tasks that you are likely to do are:
Defining parameters
Running a simulation
Plotting results
This tutorial walks you through the simplest possible version of how to do these things.
An interactive version of this notebook is available on Google Colab or Binder.
Hello world#
Let’s start with the simplest version of a Starsim model. We’ll make a version of a classic SIR model. Here’s how our code would look:
[1]:
import starsim as ss
# Define the parameters
pars = dict(
n_agents = 10_000, # Number of agents to simulate
networks = dict( # *Networks* add detail on how the agents interact with each other
type = 'random', # Here, we use a 'random' network
n_contacts = 10 # Each person has an average of 10 contacts with other people
),
diseases = dict( # *Diseases* add detail on what diseases to model
type = 'sir', # Here, we're creating an SIR disease
init_prev = 0.01, # Proportion of the population initially infected
beta = 0.05, # Probability of transmission between contacts
)
)
# Make the sim, run and plot
sim = ss.Sim(pars)
sim.run()
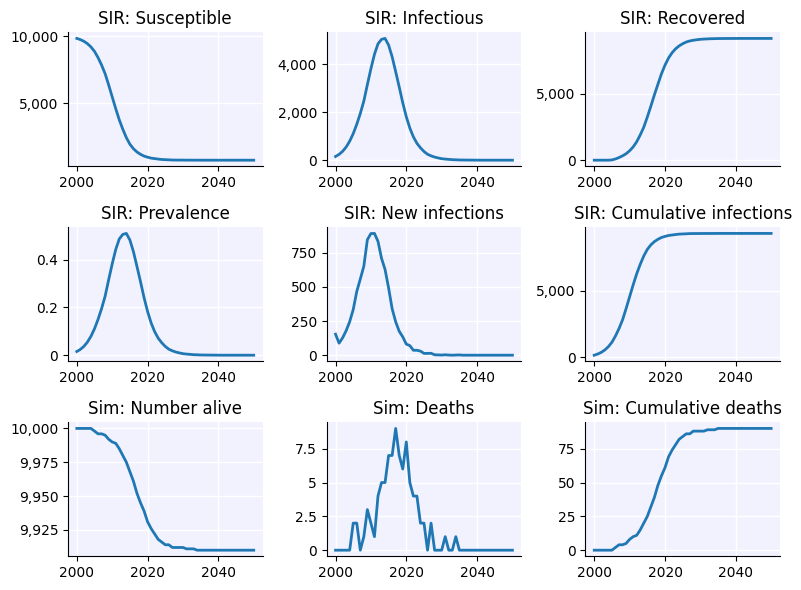
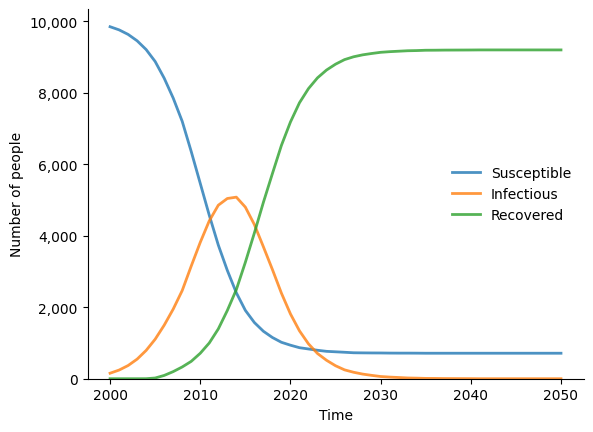
sim.plot()
sim.diseases.sir.plot()
Initializing sim with 10000 agents
Running 2000.0 ( 0/51) (0.00 s) ———————————————————— 2%
Running 2010.0 (10/51) (0.07 s) ••••———————————————— 22%
Running 2020.0 (20/51) (0.14 s) ••••••••———————————— 41%
Running 2030.0 (30/51) (0.20 s) ••••••••••••———————— 61%
Running 2040.0 (40/51) (0.25 s) ••••••••••••••••———— 80%
Running 2050.0 (50/51) (0.30 s) •••••••••••••••••••• 100%
Figure(800x600)
Figure(640x480)
As the tutorials progress we’ll show how to extend this different diseases with different transmission pathways, and how to customize the model in lots of different ways.
