Developer tutorial: Diseases#
An interactive version of this notebook is available on Google Colab or Binder.
Overview of Starsim’s disease structure#
The basic template for modeling a disease in Starsim is the Disease class. Much like sims or networks, a Disease can be customized by passing in a pars dictionary containing parameters. The Disease module does lots of different things, but three of the model fundamental are:
set_initial_states, which initializes people into states (e.g. susceptible, infected, recovered)make_new_cases, which makes new cases (e.g., by finding the susceptible contacts of all infected people and calculating the probability of transmission for each)set_prognoses, which sets the outcomes for people who get infected (e.g., by setting their date of recovery or death).
Making your own disease#
If you want to make your own disease, you could either inherit from one of the templates in diseases.py, or you could copy the examples and extend them to capture features of the disease that you want to model. For example, suppose you wanted to change the SIR model to an SEIR model (i.e., add an ‘exposed’ state where people were transmissible but did not yet have symptoms. You might hope that this would be a relatively simple change to make. Here’s how it would look:
[1]:
import starsim as ss
import matplotlib.pyplot as plt
class SEIR(ss.SIR):
def __init__(self, pars=None, *args, **kwargs):
super().__init__()
self.define_pars(
dur_exp = ss.lognorm_ex(0.5),
)
self.update_pars(pars, **kwargs)
# Additional states beyond the SIR ones
self.define_states(
ss.State('exposed', label='Exposed'),
ss.FloatArr('ti_exposed', label='TIme of exposure'),
)
return
@property
def infectious(self):
return self.infected | self.exposed
def step_state(self):
""" Make all the updates from the SIR model """
# Perform SIR updates
super().step_state()
# Additional updates: progress exposed -> infected
infected = self.exposed & (self.ti_infected <= self.ti)
self.exposed[infected] = False
self.infected[infected] = True
return
def step_die(self, uids):
super().step_die(uids)
self.exposed[uids] = False
return
def set_prognoses(self, uids, sources=None):
""" Carry out state changes associated with infection """
super().set_prognoses(uids, sources)
ti = self.ti
self.susceptible[uids] = False
self.exposed[uids] = True
self.ti_exposed[uids] = ti
# Calculate and schedule future outcomes
dur_exp = self.pars['dur_exp'].rvs(uids)
self.ti_infected[uids] = ti + dur_exp
dur_inf = self.pars['dur_inf'].rvs(uids)
will_die = self.pars['p_death'].rvs(uids)
self.ti_recovered[uids[~will_die]] = ti + dur_inf[~will_die]
self.ti_dead[uids[will_die]] = ti + dur_inf[will_die]
return
def plot(self):
""" Update the plot with the exposed compartment """
with ss.options.context(jupyter=False):
fig = super().plot()
ax = plt.gca()
res = self.results.n_exposed
ax.plot(res.timevec, res, label=res.label)
plt.legend()
return ss.return_fig(fig)
The new class includes the following main changes:
In
__init__we added the extra pars and states needed for our modelWe defined
infectiousto include both infected and exposed people - this means that we can just reuse the existing logic for how the SIR model handles transmissionWe updated
update_preandupdate_deathto include changes to theexposedstateWe rewrote
set_prognosesto include the new exposed state.
Here’s how it would look in practice:
[2]:
seir = SEIR()
sim = ss.Sim(diseases=seir, networks='random')
sim.run()
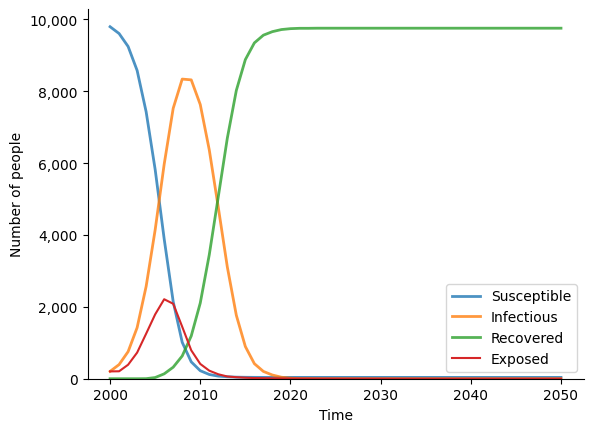
sim.plot()
sim.diseases.seir.plot()
Initializing sim with 10000 agents
Running 2000.0 ( 0/51) (0.00 s) ———————————————————— 2%
Running 2010.0 (10/51) (0.08 s) ••••———————————————— 22%
Running 2020.0 (20/51) (0.16 s) ••••••••———————————— 41%
Running 2030.0 (30/51) (0.21 s) ••••••••••••———————— 61%
Running 2040.0 (40/51) (0.27 s) ••••••••••••••••———— 80%
Running 2050.0 (50/51) (0.32 s) •••••••••••••••••••• 100%
Figure(933.333x700)
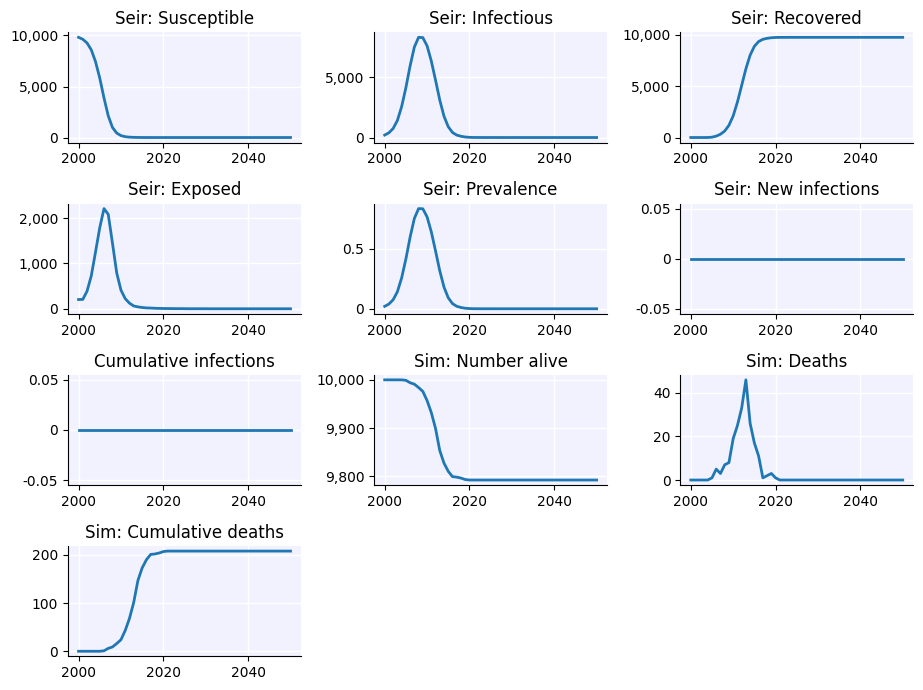
Figure(640x480)