T2 - Plotting, printing, and saving#
This tutorial provides a brief overview of options for plotting results, printing objects, and saving results.
Click here to open an interactive version of this notebook.
Global plotting configuration#
Covasim allows the user to set various options that apply to all plots. You can change the font size, default DPI, whether plots should be shown by default, etc. (for the full list, see cv.options.help()). For example, we might want higher resolution, to turn off automatic figure display, close figures after they’re rendered, and to turn off the messages that print when a simulation is running. We can do this using built-in defaults for Jupyter notebooks (and then run a sim) with:
[1]:
import covasim as cv
cv.options(jupyter=True, verbose=0) # Standard options for Jupyter notebook
sim = cv.Sim()
sim.run()
Covasim 3.1.7 (2026-02-18) — © 2020-2026 by IDM
[1]:
Sim(<no label>; 2020-03-01 to 2020-04-30; pop: 20000 random; epi: 12730⚙, 14☠)
Printing objects#
There are three levels of detail available for most objects (sims, multisims, scenarios, and people). The shortest is brief():
[2]:
sim.brief()
Sim(<no label>; 2020-03-01 to 2020-04-30; pop: 20000 random; epi: 12730⚙, 14☠)
You can get more detail with summarize():
[3]:
sim.summarize()
Simulation summary:
12,730 cumulative infections
596 cumulative reinfections
10,182 cumulative infectious
6,395 cumulative symptomatic cases
386 cumulative severe cases
114 cumulative critical cases
5,159 cumulative recoveries
14 cumulative deaths
0 cumulative tests
0 cumulative diagnoses
0 cumulative known deaths
0 cumulative quarantined people
0 cumulative isolated people
0 cumulative vaccine doses
0 cumulative vaccinated people
Finally, to show the full object, including all methods and attributes, use disp():
[4]:
sim.disp()
<covasim.sim.Sim at 0x7c3c21733700>
[<class 'covasim.sim.Sim'>, <class 'covasim.base.BaseSim'>, <class 'covasim.base.ParsObj'>, <class 'covasim.base.FlexPretty'>, <class 'sciris.sc_printing.prettyobj'>]
————————————————————————————————————————————————————————————————————————
Methods:
_brief() finalize_analyzers() plot()
_disp() finalize_interventi... plot_result()
_get_ia() get_analyzer() rescale()
brief() get_analyzers() reset_layer_pars()
calibrate() get_intervention() result_keys()
compute_doubling() get_interventions() run()
compute_fit() init_analyzers() save()
compute_gen_time() init_immunity() set_metadata()
compute_r_eff() init_infections() set_seed()
compute_results() init_interventions() shrink()
compute_states() init_people() step()
compute_summary() init_results() summarize()
compute_yield() init_variants() to_df()
copy() initialize() to_excel()
date() layer_keys() to_json()
day() load() update_pars()
disp() load_data() validate_layer_pars()
export_pars() load_population() validate_pars()
export_results() make_age_histogram() finalize()
make_transtree()
————————————————————————————————————————————————————————————————————————
Properties:
datevec npts tvec
n scaled_pop_size
————————————————————————————————————————————————————————————————————————
_default_ver: None
_legacy_trans: False
_orig_pars: {'interventions': [], 'analyzers': []}
complete: True
created: datetime.datetime(2026, 2, 19, 4, 20, 31, 816613)
data: None
datafile: None
git_info: {'covasim': {'version': '3.1.7', 'branch': 'Branch
N/A', 'hash': 'H [...]
initialized: True
label: None
pars: {'pop_size': 20000, 'pop_infected': 20, 'pop_type':
'random', 'loca [...]
people: People(n=20000; layers: a)
popdict: None
popfile: None
rescale_vec: array([1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
1., 1., 1., [...]
results: #0. 'cum_infections':
<covasim.base.Result at 0x7c3c216c0df0>
[<cla [...]
results_ready: True
simfile: 'covasim.sim'
summary: #0. 'cum_infections': 12730.0
#1. 'cum_reinfections': 596.0 [...]
t: 60
version: '3.1.7'
————————————————————————————————————————————————————————————————————————
Plotting options#
While a sim can be plotted using default settings simply by sim.plot(), this is just a small fraction of what’s available. First, note that results can be plotted directly using e.g. Matplotlib. You can see what quantities are available for plotting with sim.results.keys() (remember, it’s just a dict). A simple example of plotting using Matplotlib is:
[5]:
import pylab as pl # Shortcut for import matplotlib.pyplot as plt
pl.plot(sim.results['date'], sim.results['new_infections'])
[5]:
[<matplotlib.lines.Line2D at 0x7c3be5b08d60>]
However, as you can see, this isn’t ideal since the default formatting is…not great. (Also, note that each result is a Result object, not a simple Numpy array; like a pandas dataframe, you can get the array of values directly via e.g. sim.results.new_infections.values.)
An alternative, if you only want to plot a single result, such as new infections, is to use the plot_result() method:
[6]:
sim.plot_result('new_infections')
You can also select one or more quantities to plot with the first (to_plot) argument, e.g.
[7]:
sim.plot(to_plot=['new_infections', 'cum_infections'])
Another useful option is to plot an overview of everything in a simulation. We can do this with the to_plot='overview' argument. It’s quite a lot of information so we might also want to make a larger figure for it, which we can do by passing additional arguments (which, if recognized, are passed to pl.figure()). We can also change the date format between 'sciris' (the default), 'concise' (better for cramped plots), and 'matplotlib' (Matplotlib’s default):
[8]:
sim.plot('overview', n_cols=5, figsize=(20,20), dateformat='concise', dpi=50) # NB: dateformat='concise' is already the default for >2 columns
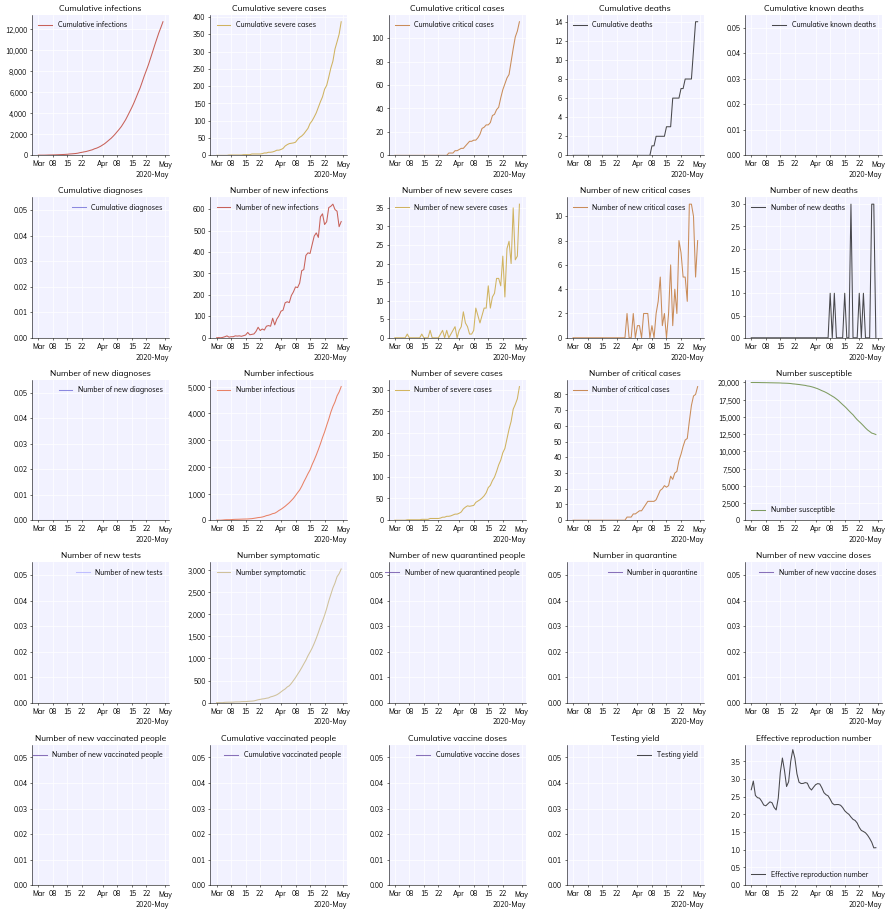
While we can save this figure using Matplotlib’s built-in savefig(), if we use Covasim’s cv.savefig() we get a couple advantages:
[9]:
cv.savefig(filename='my-fig.png')
[9]:
'my-fig.png'
<Figure size 640x480 with 0 Axes>
First, it saves the figure at higher resolution by default (which you can adjust with the dpi argument). But second, it stores information about the code that was used to generate the figure as metadata, which can be loaded later. Made an awesome plot but can’t remember even what script you ran to generate it, much less what version of the code? You’ll never have to worry about that again.
[10]:
cv.get_png_metadata('my-fig.png')
Covasim version: 3.1.7
Covasim branch: Branch N/A
Covasim hash: Hash N/A
Covasim date: Date N/A
Covasim caller branch: Branch N/A
Covasim caller hash: Hash N/A
Covasim caller date: Date N/A
Covasim caller filename: /home/docs/checkouts/readthedocs.org/user_builds/institute-for-disease-modeling-covasim/envs/latest/lib/python3.9/site-packages/covasim/misc.py
Covasim current time: 2026-Feb-19 04:20:36
Covasim calling file: /tmp/ipykernel_2382/1033588508.py
Customizing plots#
We saw above how to set default plot configuration options for Jupyter. Covasim provides a lot of flexibility in customizing the appearance of plots as well. There are three different levels at which you can set plotting options: global, just for Covasim, or just for the current plot. To give an example with changing the figure DPI:
Change the setting globally (for both Covasim and Matplotlib):
sc.options(dpi=150)orpl.rc('figure', dpi=150)(wherescisimport sciris as sc)Change for Covasim plots, but not for Matplotlib plots:
cv.options(dpi=150)Change for the current Covasim plot, but not other Covasim plots:
sim.plot(dpi=150)
The easiest way to change the style of Covasim plots is with the style argument. For example, to plot using a built-in Matplotlib style would simply be:
[11]:
sim.plot(style='ggplot')
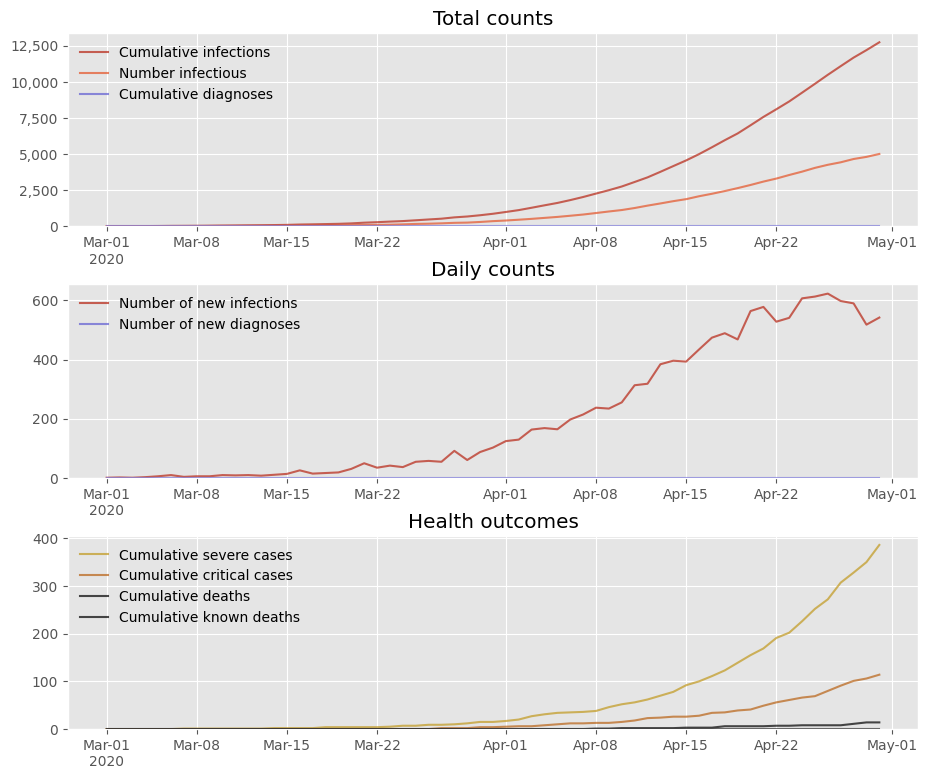
In addition to the default style ('covasim'), there is also a “simple” style. You can combine built-in styles with additional overrides, including any valid Matplotlib commands:
[12]:
sim.plot(style='simple', font='Rosario', legend_args={'frameon':True}, style_args={'ytick.direction':'in'})
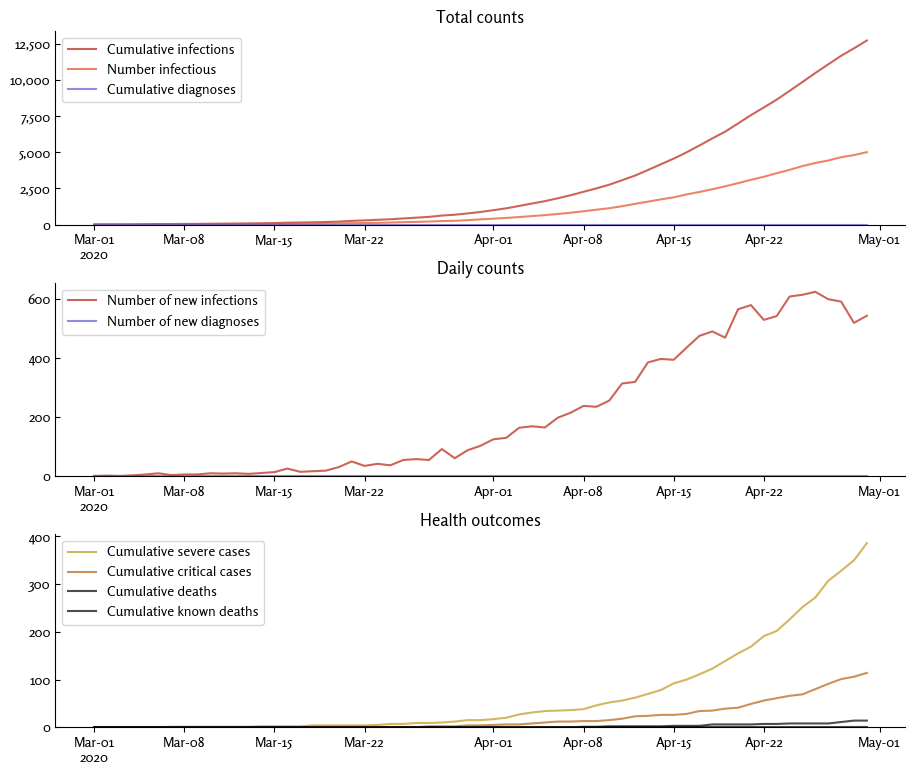
(Note: Covasim comes bundled with two fonts, Mulish (sans-serif, the default) and Rosario (serif).)
Although most style handling is done automatically, you can also use it yourself in a with block, e.g.:
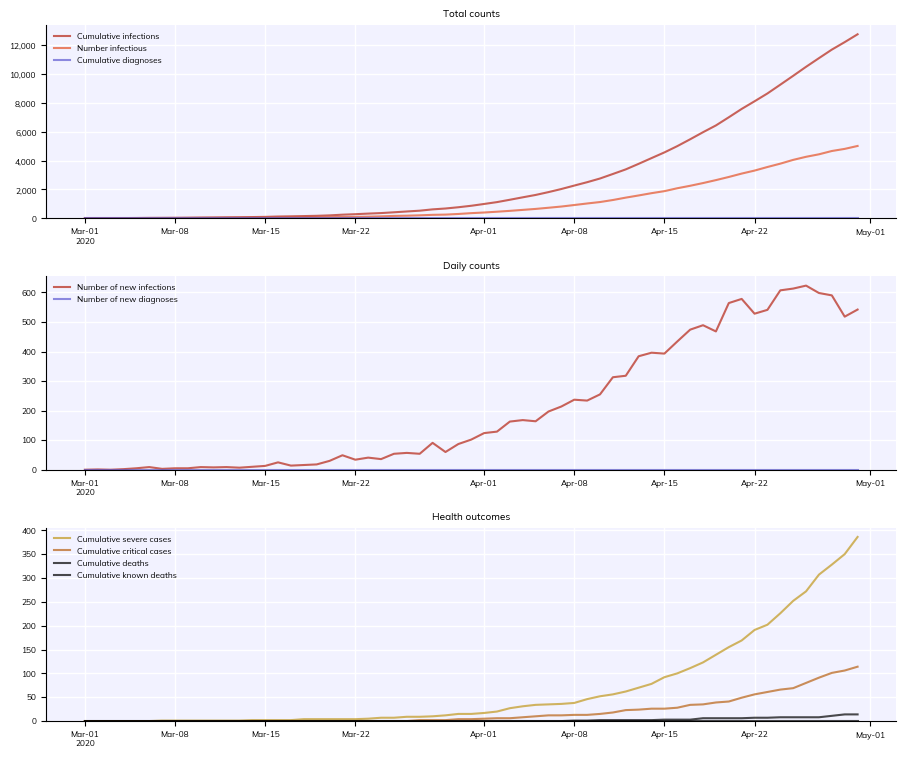
[13]:
with cv.options.with_style(fontsize=6):
sim.plot() # This will have 6 point font
pl.figure(); pl.plot(pl.rand(20), 'o') # So will this
Saving options#
Saving sims is also pretty simple. The simplest way to save is simply
[14]:
sim.save('my-awesome-sim.sim')
[14]:
'/home/docs/checkouts/readthedocs.org/user_builds/institute-for-disease-modeling-covasim/checkouts/latest/docs/tutorials/my-awesome-sim.sim'
Technically, this saves as a gzipped pickle file (via sc.saveobj() using the Sciris library). By default this does not save the people in the sim since they are very large (and since, if the random seed is saved, they can usually be regenerated). If you want to save the people as well, you can use the keep_people argument. For example, here’s what it would look like to create a sim, run it halfway, save it, load it, change the overall transmissibility (beta), and finish running it:
[15]:
sim_orig = cv.Sim(start_day='2020-04-01', end_day='2020-06-01', label='Load & save example')
sim_orig.run(until='2020-05-01')
sim_orig.save('my-half-finished-sim.sim') # Note: Covasim always saves the people if the sim isn't finished running yet
sim = cv.load('my-half-finished-sim.sim')
sim['beta'] *= 0.3
sim.run()
sim.plot(['new_infections', 'n_infectious', 'cum_infections'])
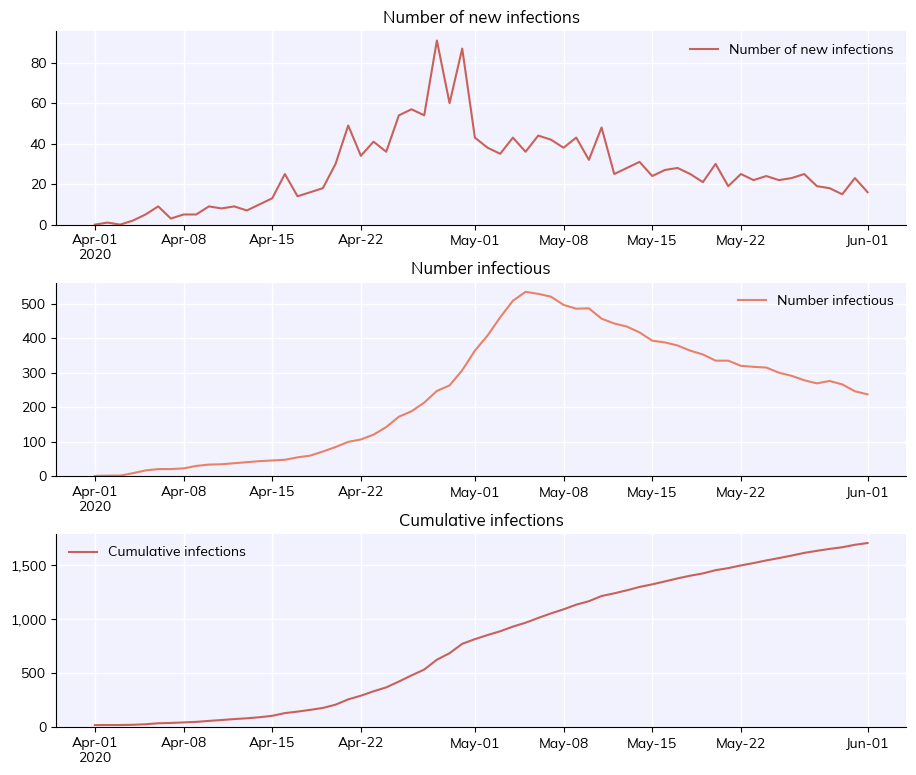
Aside from saving the entire simulation, there are other export options available. You can export the results and parameters to a JSON file (using sim.to_json()), but probably the most useful is to export the results to an Excel workbook, where they can easily be stored and processed with e.g. Pandas:
[16]:
import pandas as pd
sim.to_excel('my-sim.xlsx')
df = pd.read_excel('my-sim.xlsx')
print(df)
Object saved to /home/docs/checkouts/readthedocs.org/user_builds/institute-for-disease-modeling-covasim/checkouts/latest/docs/tutorials/my-sim.xlsx.
date t cum_infections cum_reinfections cum_infectious \
0 2020-04-01 0 20 0 0
1 2020-04-02 1 21 0 1
2 2020-04-03 2 21 0 1
3 2020-04-04 3 23 0 8
4 2020-04-05 4 28 0 16
.. ... .. ... ... ...
57 2020-05-28 57 1632 25 1527
58 2020-05-29 58 1650 25 1561
59 2020-05-30 59 1665 25 1579
60 2020-05-31 60 1688 25 1596
61 2020-06-01 61 1704 26 1615
cum_symptomatic cum_severe cum_critical cum_recoveries cum_deaths \
0 0 0 0 0 0
1 0 0 0 0 0
2 0 0 0 0 0
3 0 0 0 0 0
4 7 0 0 0 0
.. ... ... ... ... ...
57 1053 92 31 1251 7
58 1069 94 31 1278 7
59 1089 98 31 1306 7
60 1096 99 32 1341 9
61 1109 101 32 1369 9
... prevalence incidence r_eff doubling_time test_yield \
0 ... 0.001000 0.000000 2.766969 NaN 0
1 ... 0.001050 0.000050 3.018511 NaN 0
2 ... 0.001050 0.000000 2.602426 NaN 0
3 ... 0.001150 0.000100 2.537009 14.878453 0
4 ... 0.001400 0.000250 2.520561 7.228263 0
.. ... ... ... ... ... ...
57 ... 0.018707 0.000968 0.738915 30.000000 0
58 ... 0.018256 0.000917 0.678174 30.000000 0
59 ... 0.017606 0.000764 0.696037 30.000000 0
60 ... 0.016908 0.001170 0.936910 30.000000 0
61 ... 0.016307 0.000814 0.676514 30.000000 0
rel_test_yield frac_vaccinated pop_nabs pop_protection \
0 0 0 0.000258 0.000000
1 0 0 0.000518 0.000000
2 0 0 0.000778 0.000000
3 0 0 0.001047 0.000000
4 0 0 0.001348 0.000000
.. ... ... ... ...
57 0 0 0.486243 0.050087
58 0 0 0.489911 0.051117
59 0 0 0.493096 0.052214
60 0 0 0.495393 0.053432
61 0 0 0.497245 0.054493
pop_symp_protection
0 0.000000
1 0.000000
2 0.000000
3 0.000000
4 0.000000
.. ...
57 0.020291
58 0.020736
59 0.021199
60 0.021766
61 0.022223
[62 rows x 60 columns]
