T7 - Calibration#
Tutorial 2 demonstrated how to run the model and plot the outputs. But it’s entirely possible that the model outputs won’t look like the data for the country that you wish to model. The default parameter values included in HPVsim are intended as points of departure to be iteratively refined via calibration. The process of model calibration involves finding the model parameters that are the most likely explanation for the observed data. This tutorial gives an introduction to the Fit object and some recipes for optimization approaches.
Click here to open an interactive version of this notebook.
Data types supported by HPVsim#
Data on HPV and cervical disease comes in many different formats. When using HPVsim, the goal is typically to produce population-level estimates of epidemic outputs like: - age-specific incidence of cancer or high-grade lesions in one or more years; - number of cases of cancer or high-grade lesions reported in one or more years; - HPV prevalence over time; - lifetime incidence of HPV; - the distribution of genotypes in detected cases of cancer/high-grade lesions; - sexual behavior metrics like the average age at first marriage, duration of relationships, or number of lifetime partners.
After running HPVsim, estimates all of these variables are included within the results dictionary. To plot them alongside data, the easiest method is to use the Calibration object.
The Calibration object#
Calibration objects contain the following ingredients: - an hpv.Sim() instance with details of the model configuration; - two lists of parameters to vary, one for parameters that vary by genotype and one for those that don’t; - dataframes that hold the calibration targets, which are typically added as csv files; - a list of any additional results to plot; - settings that are passed to the Optuna package[LINK], an open source hyperparameter optimization framework that automates calibration
for HPVsim.
We have included Optuna as a built-in calibration option as we have found that it works reasonably well, but it is also possible to use other methods; we will discuss this a little further down.
The example below illustrates the general idea of calibration, and can be adapted for different use cases:
[1]:
# Import HPVsim
import hpvsim as hpv
# Configure a simulation with some parameters
pars = dict(n_agents=10e3, start=1980, end=2020, dt=0.25, location='nigeria')
sim = hpv.Sim(pars)
# Specify some parameters to adjust during calibration.
# The parameters in the calib_pars dictionary don't vary by genotype,
# whereas those in the genotype_pars dictionary do. Both kinds are
# given in the order [best, lower_bound, upper_bound].
calib_pars = dict(
beta=[0.05, 0.010, 0.20],
)
genotype_pars = dict(
hpv16=dict(
cin_fn=dict(k=[0.5, 0.2, 1.0]),
dur_cin=dict(par1=[6, 4, 12])
),
hpv18=dict(
cin_fn=dict(k=[0.5, 0.2, 1.0]),
dur_cin=dict(par1=[6, 4, 12])
)
)
# List the datafiles that contain data that we wish to compare the model to:
datafiles=['nigeria_cancer_cases.csv',
'nigeria_cancer_types.csv']
# List extra results that we don't have data on, but wish to include in the
# calibration object so we can plot them.
results_to_plot = ['cancer_incidence', 'asr_cancer_incidence']
# Create the calibration object, run it, and plot the results
calib = hpv.Calibration(
sim,
calib_pars=calib_pars,
genotype_pars=genotype_pars,
extra_sim_result_keys=results_to_plot,
datafiles=datafiles,
total_trials=3, n_workers=1
)
calib.calibrate(die=True)
calib.plot(res_to_plot=4);
HPVsim 2.1.0 (2025-03-25) — © 2023-2025 by IDM
Loading location-specific demographic data for "nigeria"
Initializing sim with 10000 agents
Loading location-specific data for "nigeria"
/home/docs/checkouts/readthedocs.org/user_builds/institute-for-disease-modeling-hpvsim/envs/latest/lib/python3.9/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
[I 2025-07-15 18:12:02,224] A new study created in RDB with name: hpvsim_calibration
Could not delete study, skipping...
'Record does not exist.'
Removed existing calibration hpvsim_calibration.db
Initializing sim (resetting people) with 10000 agents
Loading location-specific data for "nigeria"
Running 1980.0 ( 0/164) (0.00 s) ———————————————————— 1%
Running 1982.5 (10/164) (0.12 s) •——————————————————— 7%
Running 1985.0 (20/164) (0.24 s) ••—————————————————— 13%
Running 1987.5 (30/164) (0.37 s) •••————————————————— 19%
Running 1990.0 (40/164) (0.50 s) •••••——————————————— 25%
Running 1992.5 (50/164) (0.63 s) ••••••—————————————— 31%
Running 1995.0 (60/164) (0.76 s) •••••••————————————— 37%
Running 1997.5 (70/164) (0.91 s) ••••••••———————————— 43%
Running 2000.0 (80/164) (1.05 s) •••••••••——————————— 49%
Running 2002.5 (90/164) (1.20 s) •••••••••••————————— 55%
Running 2005.0 (100/164) (1.35 s) ••••••••••••———————— 62%
Running 2007.5 (110/164) (1.52 s) •••••••••••••——————— 68%
Running 2010.0 (120/164) (1.69 s) ••••••••••••••—————— 74%
Running 2012.5 (130/164) (1.86 s) •••••••••••••••————— 80%
Running 2015.0 (140/164) (2.03 s) •••••••••••••••••——— 86%
Running 2017.5 (150/164) (2.21 s) ••••••••••••••••••—— 92%
[I 2025-07-15 18:12:04,826] Trial 0 finished with value: 56.66500029548874 and parameters: {'hpv16_cin_fn_k': 0.7227064907857057, 'hpv16_dur_cin_par1': 9.843036612366268, 'hpv18_cin_fn_k': 0.406387450444163, 'hpv18_dur_cin_par1': 4.602628575724818, 'beta': 0.1702989440033317}. Best is trial 0 with value: 56.66500029548874.
Running 2020.0 (160/164) (2.40 s) •••••••••••••••••••— 98%
Simulation summary:
767,261,820 total HPV infections
2,092,462 total cancers
1,148,603 total cancer deaths
5.68 mean HPV prevalence (%)
72.97 mean cancer incidence (per 100k)
35.23 mean age of infection (years)
42.92 mean age of cancer (years)
50.36 mean age of cancer death (years)
Initializing sim (resetting people) with 10000 agents
Loading location-specific data for "nigeria"
Running 1980.0 ( 0/164) (0.00 s) ———————————————————— 1%
Running 1982.5 (10/164) (0.13 s) •——————————————————— 7%
Running 1985.0 (20/164) (0.25 s) ••—————————————————— 13%
Running 1987.5 (30/164) (0.38 s) •••————————————————— 19%
Running 1990.0 (40/164) (0.51 s) •••••——————————————— 25%
Running 1992.5 (50/164) (0.64 s) ••••••—————————————— 31%
Running 1995.0 (60/164) (0.78 s) •••••••————————————— 37%
Running 1997.5 (70/164) (0.92 s) ••••••••———————————— 43%
Running 2000.0 (80/164) (1.07 s) •••••••••——————————— 49%
Running 2002.5 (90/164) (1.21 s) •••••••••••————————— 55%
Running 2005.0 (100/164) (1.36 s) ••••••••••••———————— 62%
Running 2007.5 (110/164) (1.52 s) •••••••••••••——————— 68%
Running 2010.0 (120/164) (1.68 s) ••••••••••••••—————— 74%
Running 2012.5 (130/164) (1.85 s) •••••••••••••••————— 80%
Running 2015.0 (140/164) (2.02 s) •••••••••••••••••——— 86%
Running 2017.5 (150/164) (2.19 s) ••••••••••••••••••—— 92%
[I 2025-07-15 18:12:07,496] Trial 1 finished with value: 19.887359624296067 and parameters: {'hpv16_cin_fn_k': 0.4967061326626412, 'hpv16_dur_cin_par1': 5.987012173843757, 'hpv18_cin_fn_k': 0.44978075574410276, 'hpv18_dur_cin_par1': 9.858382171578015, 'beta': 0.12385198814136596}. Best is trial 1 with value: 19.887359624296067.
Running 2020.0 (160/164) (2.38 s) •••••••••••••••••••— 98%
Simulation summary:
612,964,927 total HPV infections
1,072,368 total cancers
622,947 total cancer deaths
4.63 mean HPV prevalence (%)
37.57 mean cancer incidence (per 100k)
35.09 mean age of infection (years)
43.09 mean age of cancer (years)
47.72 mean age of cancer death (years)
Initializing sim (resetting people) with 10000 agents
Loading location-specific data for "nigeria"
Running 1980.0 ( 0/164) (0.00 s) ———————————————————— 1%
Running 1982.5 (10/164) (0.12 s) •——————————————————— 7%
Running 1985.0 (20/164) (0.23 s) ••—————————————————— 13%
Running 1987.5 (30/164) (0.35 s) •••————————————————— 19%
Running 1990.0 (40/164) (0.47 s) •••••——————————————— 25%
Running 1992.5 (50/164) (0.59 s) ••••••—————————————— 31%
Running 1995.0 (60/164) (0.71 s) •••••••————————————— 37%
Running 1997.5 (70/164) (0.84 s) ••••••••———————————— 43%
Running 2000.0 (80/164) (0.97 s) •••••••••——————————— 49%
Running 2002.5 (90/164) (1.11 s) •••••••••••————————— 55%
Running 2005.0 (100/164) (1.24 s) ••••••••••••———————— 62%
Running 2007.5 (110/164) (1.39 s) •••••••••••••——————— 68%
Running 2010.0 (120/164) (1.53 s) ••••••••••••••—————— 74%
Running 2012.5 (130/164) (1.69 s) •••••••••••••••————— 80%
Running 2015.0 (140/164) (1.85 s) •••••••••••••••••——— 86%
Running 2017.5 (150/164) (2.00 s) ••••••••••••••••••—— 92%
Running 2020.0 (160/164) (2.17 s) •••••••••••••••••••— 98%
[I 2025-07-15 18:12:09,844] Trial 2 finished with value: 9.117852334524127 and parameters: {'hpv16_cin_fn_k': 0.29195546311812626, 'hpv16_dur_cin_par1': 5.740747335456538, 'hpv18_cin_fn_k': 0.659714199320415, 'hpv18_dur_cin_par1': 11.90879850298108, 'beta': 0.05300689413532489}. Best is trial 2 with value: 9.117852334524127.
Simulation summary:
249,002,936 total HPV infections
683,208 total cancers
422,558 total cancer deaths
2.39 mean HPV prevalence (%)
24.56 mean cancer incidence (per 100k)
39.48 mean age of infection (years)
50.09 mean age of cancer (years)
53.88 mean age of cancer death (years)
Loading saved results...
Removed temporary file tmp_calibration_00000.obj
Loaded trial 0
Removed temporary file tmp_calibration_00001.obj
Loaded trial 1
Removed temporary file tmp_calibration_00002.obj
Loaded trial 2
Making results structure...
Processed 3 trials; 0 failed
Deleted study hpvsim_calibration in sqlite:///hpvsim_calibration.db
Removed existing calibration hpvsim_calibration.db
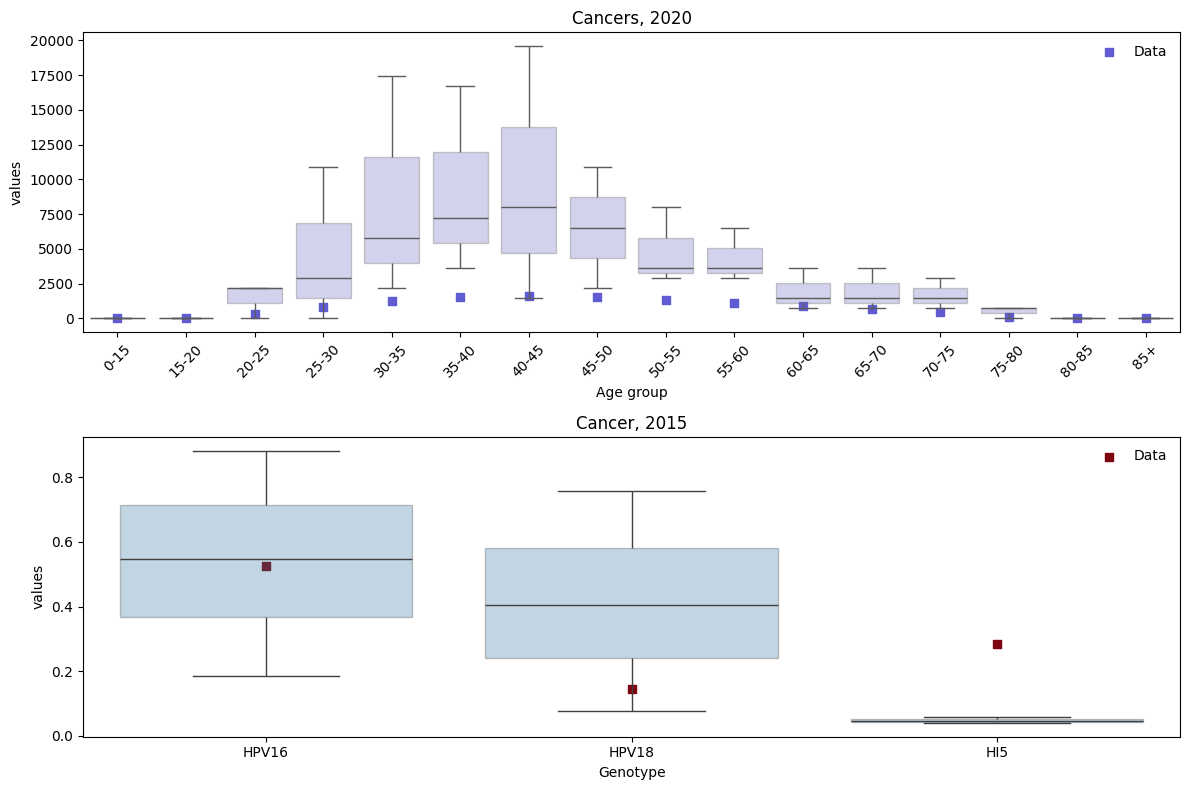
This isn’t a great fit yet! In general, it will probably be necessary to run many more trials that the 3 we ran here. Moreover, careful consideration should be given to the parameters that you want to adjust during calibration. In HPVsim we have taken the approach that any parameter can be adjusted. As we learn more about which parameters make most sense to calibrate, we will add details here. We would also enourage users to share their experiences with calibration and parameter searches.
