T5 - Using interventions#
Interventions are one of the most critical parts of HPVsim. This tutorial shows how to implement standard interventions, as well as how to define your own custom interventions.
Click here to open an interactive version of this notebook.
Products and interventions#
HPVsim contains products, which can be thought of as the actual test, diagnostic, treatment, or vaccine product being used, as well as interventions, which are responsible for delivering the products to the population. Information about the default products included with HPVsim (e.g. attributes like test positivity and efficacy) are available in the hpvsim/data/products_*.csv files. Specifically:
Screening products (VIA, Pap smears, liquid-based cytology, HPV testing, and HPV16/18 testing):
hpvsim/data/products_dx.csvTreatment products (ablation, excision, and therapeutic vaccines):
hpvsim/data/products_tx.csvProphylactic vaccine products (bivalent, quadrivalent, nonavalent):
hpvsim/data/products_vx.csv
It’s also possible to make a custom product, e.g.
[1]:
import hpvsim as hpv
import pandas as pd
my_treatment_data = pd.DataFrame({'name':'new_tx', 'state':['precin','cin1','cin2','cin3','cancerous'],'genotype':'all','efficacy':[.2,.3,.3,.4,.4]})
my_treatment = hpv.tx(df=my_treatment_data)
HPVsim 2.1.0 (2025-03-25) — © 2023-2025 by IDM
Custom products can be made for diagnostics and vaccination in the same way. The efficacy of some products varies by genotype, in which case efficacy values for each genotype can be entered as separate dataframe rows.
Most of the time, it isn’t necessary to create your own products if you just want to use one of the standard options. When setting up screening, triage, or treatment interventions, it’s possible to pass in a string that will create a standard default product.
Screening and treatment interventions#
Screening and treatment is implemented in HPVsim as a flexible set of interventions that can be mixed and matched. By specifying how each of the components link together, it’s possible to create quite complex algorithms. This is illustrated in the following example:
[2]:
# Define a series of interventions to screen, triage, assign treatment, and administer treatment
prob = 0.6
screen = hpv.routine_screening(start_year=2015, prob=prob, product='via', label='screen') # Routine screening
to_triage = lambda sim: sim.get_intervention('screen').outcomes['positive'] # Define who's eligible for triage
triage = hpv.routine_triage(eligibility=to_triage, prob=prob, product='hpv', label='triage') # Triage people
to_treat = lambda sim: sim.get_intervention('triage').outcomes['positive'] # Define who's eligible to be assigned treatment
assign_tx = hpv.routine_triage(eligibility=to_treat, prob=prob, product='tx_assigner', label='assign_tx') # Assign treatment
to_ablate = lambda sim: sim.get_intervention('assign_tx').outcomes['ablation'] # Define who's eligible for ablation treatment
ablation = hpv.treat_num(eligibility=to_ablate, prob=prob, product='ablation') # Administer ablation
to_excise = lambda sim: sim.get_intervention('assign_tx').outcomes['excision'] # Define who's eligible for excision
excision = hpv.treat_delay(eligibility=to_excise, prob=prob, product='excision') # Administer excision
# Define the parameters
pars = dict(
n_agents = 20e3, # Population size
n_years = 35, # Number of years to simulate
verbose = 0, # Don't print details of the run
rand_seed = 2, # Set a non-default seed
genotypes = [16, 18], # Include the two genotypes of greatest general interest
)
# Create the sim with and without interventions
orig_sim = hpv.Sim(pars, label='Baseline')
sim = hpv.Sim(pars, interventions = [screen, triage, assign_tx, ablation, excision], label='With screen & treat')
# Run and plot
msim = hpv.parallel(orig_sim, sim)
msim.plot();
Loading location-specific demographic data for "nigeria"
Loading location-specific demographic data for "nigeria"
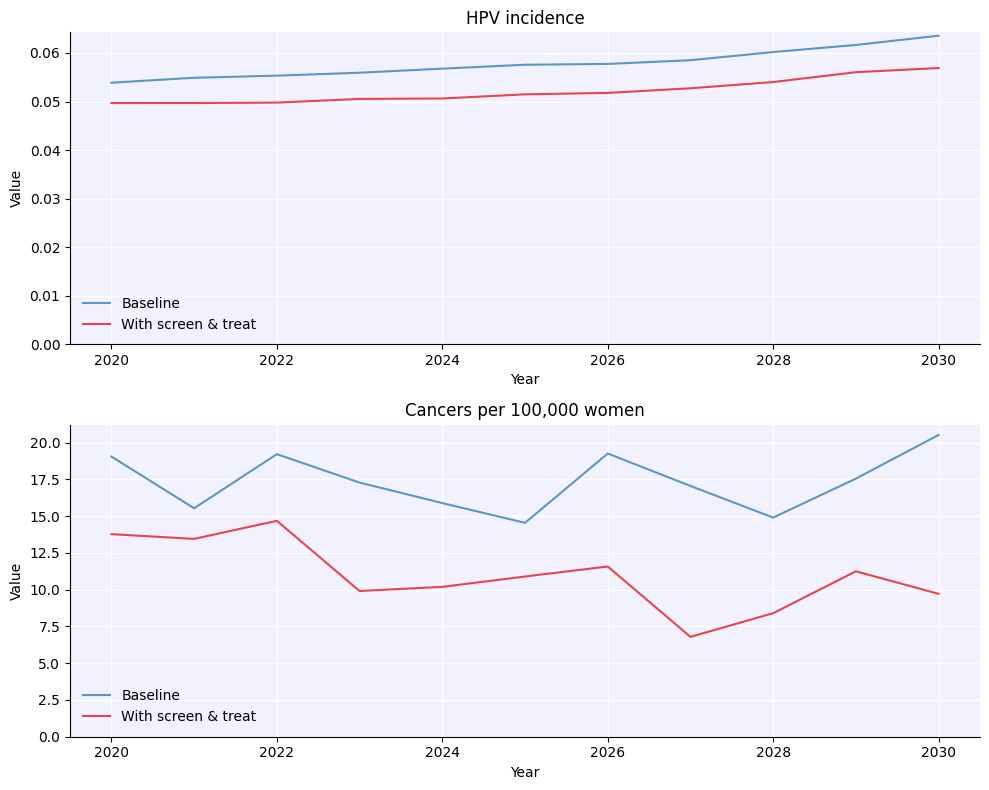
A few things to note here:
By default, interventions are shown with vertical dashed lines. You can turn this off by passing
do_plot=Falseto the intervention.Note that like other “parameters”, you can either pass interventions to the sim directly or as part of the pars dictionary; the examples below illustrate these options.
Several of the interventions above are defined as routine interventions, e.g.
hpv.routine_screening()andhpv.routine_triage(). In general, most interventions exist as both routine and campaign versions. The difference between the two comes down to how the dates are interpreted:hpv.routine_screening(start=2020, end=2030, prob=0.2)implies that the intervention will be in place each year between 2020-2030;hpv.campaign_screening(years=[2020,2030], prob=0.2)implies that the intervention will be delivered twice: once in 2020 and once in 2030.
The script examples/t05_screen_algorithms.py shows how to set up each of the seven algorithms recommended by in the WHO’s guidelined for screening and treatment of cervical pre-cancer lesions (see https://www.ncbi.nlm.nih.gov/books/NBK572308/).
Prophylactic vaccination#
Prophylactic vaccination within HPVsim simply targets a vaccine product towards a subset of the population.
[3]:
vx = hpv.routine_vx(prob=prob, start_year=2015, age_range=[9,10], product='bivalent')
# Create the sim with and without interventions
orig_sim = hpv.Sim(pars, label='Baseline')
sim = hpv.Sim(pars, interventions = vx, label='With vaccination')
# Run and plot
msim = hpv.parallel(orig_sim, sim)
msim.plot();
Loading location-specific demographic data for "nigeria"
Loading location-specific demographic data for "nigeria"
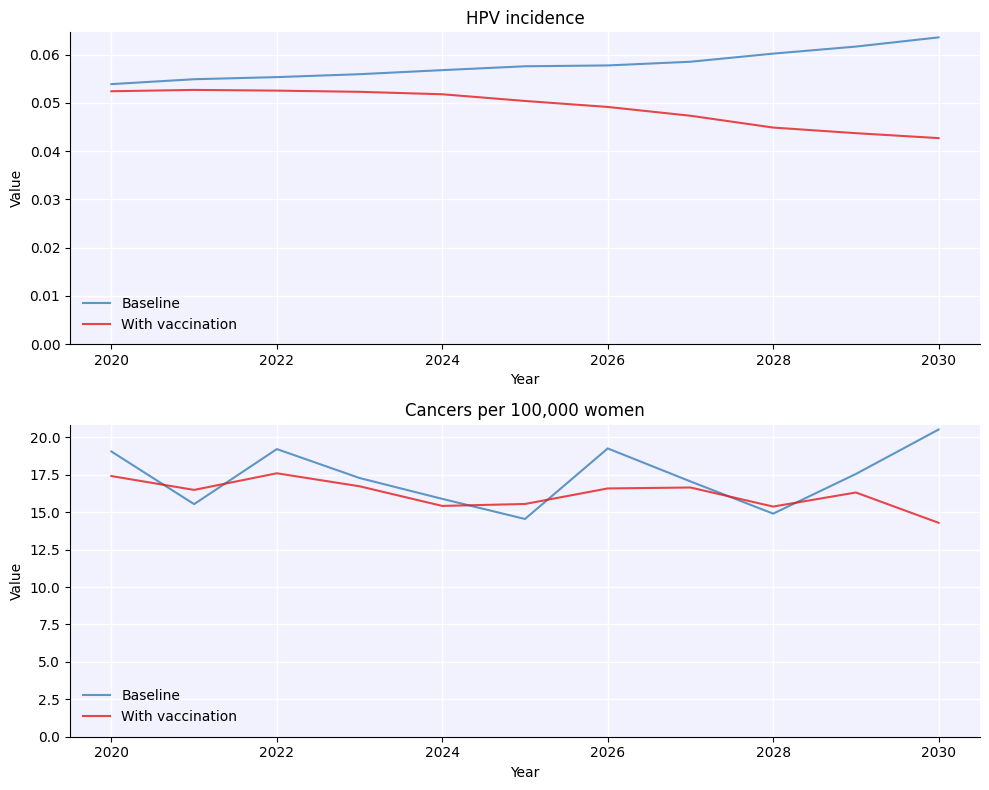
Note that probabilities passed to interventions are annual probabilities, not total.
Therapeutic vaccination#
Therapeutic vaccination can be included in a few different formats/use cases:
As a part of a screen & treat algorithm
As a mass vaccination without screening.
The following examples illustrate this:
[4]:
import numpy as np
import hpvsim as hpv
# Define mass therapeutic vaccination:
campaign_txvx_dose1 = hpv.campaign_txvx(prob = 0.9, years = 2015, age_range = [30,50], product = 'txvx1', label = 'campaign txvx')
second_dose_eligible = lambda sim: (sim.people.txvx_doses == 1) | (sim.t > (sim.people.date_tx_vaccinated + 0.5 / sim['dt']))
campaign_txvx_dose2 = hpv.campaign_txvx(prob = 0.7, years=[2015,2016], age_range=[30, 70], product = 'txvx2', eligibility = second_dose_eligible, label = 'campaign txvx 2nd dose')
routine_txvx_dose1 = hpv.routine_txvx(prob = 0.9, start_year = 2016, age_range = [30,31], product = 'txvx2',label = 'routine txvx')
second_dose_eligible = lambda sim: (sim.people.txvx_doses == 1) | (sim.t > (sim.people.date_tx_vaccinated + 0.5 / sim['dt']))
routine_txvx_dose2 = hpv.routine_txvx(prob = 0.8, start_year = 2016, age_range = [30,31], product = 'txvx1', eligibility=second_dose_eligible, label = 'routine txvx 2nd dose')
mass_vx_intvs = [campaign_txvx_dose1, campaign_txvx_dose2, routine_txvx_dose1, routine_txvx_dose2]
for intv in mass_vx_intvs: intv.do_plot=False
# Define therapeutic vaccination within screen and treat
campaign_txvx_dose1 = hpv.campaign_txvx(prob = 0.9, years = 2015, age_range = [30,50], product = 'txvx1', label = 'campaign txvx')
second_dose_eligible = lambda sim: (sim.people.txvx_doses == 1) | (sim.t > (sim.people.date_tx_vaccinated + 0.5 / sim['dt']))
campaign_txvx_dose2 = hpv.campaign_txvx(prob = 0.7, years=[2015,2016], age_range=[30, 70], product = 'txvx2', eligibility = second_dose_eligible, label = 'campaign txvx 2nd dose')
routine_txvx_dose1 = hpv.routine_txvx(prob = 0.9, start_year = 2016, age_range = [30,31], product = 'txvx2',label = 'routine txvx')
second_dose_eligible = lambda sim: (sim.people.txvx_doses == 1) | (sim.t > (sim.people.date_tx_vaccinated + 0.5 / sim['dt']))
routine_txvx_dose2 = hpv.routine_txvx(prob = 0.8, start_year = 2016, age_range = [30,31], product = 'txvx1', eligibility=second_dose_eligible, label = 'routine txvx 2nd dose')
mass_vx_intvs = [campaign_txvx_dose1, campaign_txvx_dose2, routine_txvx_dose1, routine_txvx_dose2]
for intv in mass_vx_intvs: intv.do_plot=False
# Screen, triage, assign treatment, treat
screen_eligible = lambda sim: np.isnan(sim.people.date_screened) | (sim.t > (sim.people.date_screened + 5 / sim['dt']))
routine_screen = hpv.routine_screening(start_year=2016, product='hpv', prob=0.1, eligibility=screen_eligible, age_range=[30, 50], label='routine screening')
screened_pos = lambda sim: sim.get_intervention('routine screening').outcomes['positive'] # Get those who screen positive
pos_screen_assesser = hpv.routine_triage(start_year=2016, product = 'txvx_assigner', prob = 1.0, annual_prob=False, eligibility = screened_pos, label = 'txvx assigner') # Offer TxVx or refer them for further testing
txvx_eligible = lambda sim: sim.get_intervention('txvx assigner').outcomes['txvx'] # Get people who've been classified as txvx eligible based on the positive screen assessment
deliver_txvx = hpv.linked_txvx(prob = 0.8, product = 'txvx1', eligibility = txvx_eligible, label = 'txvx') # Deliver txvx to them
screen_vx_intv = [routine_screen, pos_screen_assesser, deliver_txvx]
for intv in screen_vx_intv: intv.do_plot=False
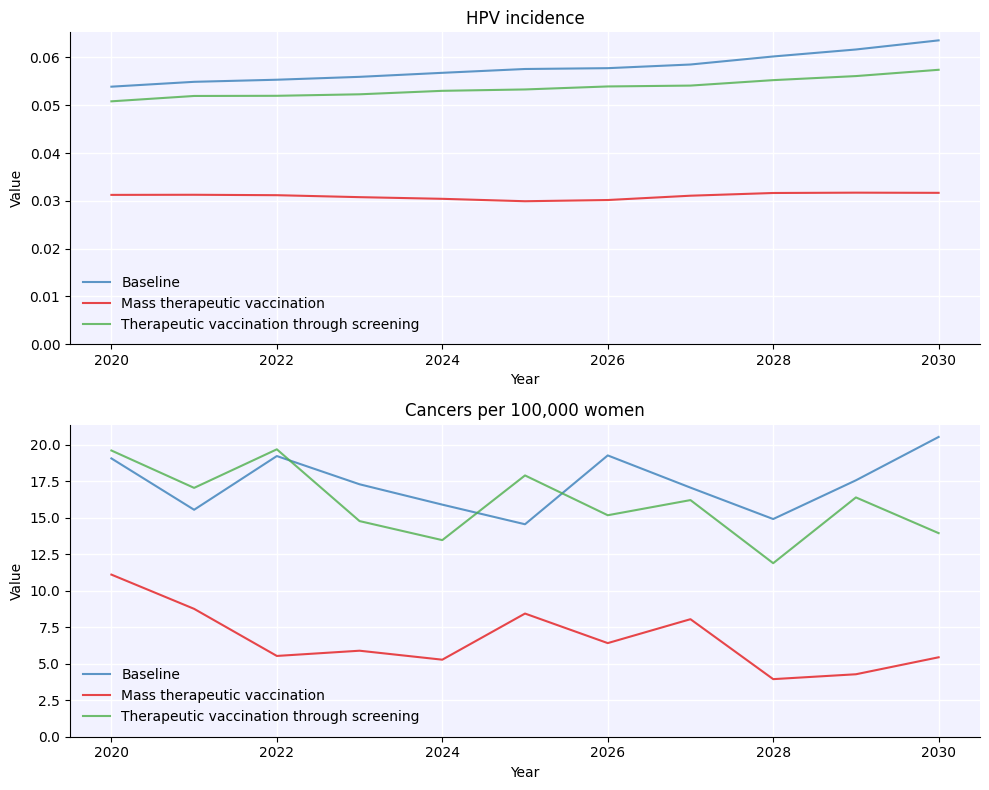
sim0 = hpv.Sim(pars=pars, label='Baseline')
sim1 = hpv.Sim(pars=pars, interventions=mass_vx_intvs, label='Mass therapeutic vaccination')
sim2 = hpv.Sim(pars=pars, interventions=screen_vx_intv, label='Therapeutic vaccination through screening')
# Run and plot
msim = hpv.parallel(sim0, sim1, sim2)
msim.plot();
Loading location-specific demographic data for "nigeria"
Loading location-specific demographic data for "nigeria"
Loading location-specific demographic data for "nigeria"