T4 - People, populations, and networks#
This tutorial gives a brief introduction to people, populations, and contact layers.
Click here to open an interactive version of this notebook.
Demographic data#
HPVsim includes pre-downloaded demographic data for almost all countries, including:
Population sizes from 1950-2100 from the UN’s World Population Projections;
Birth rates from 1950-2100 from the World Bank;
Age- and sex-specific mortality rates from 1950-2100 from the UN’s World Population Projections.
As we saw in Tutorial 1, you can load these data simply by using the location parameter. You can show a list of all available locations with hpv.data.show_locations().
People and contact network layers#
Agents in HPVsim are contained in an object called People, which contains all of the agents’ properties, as well as methods for changing them from one state to another (e.g., from susceptible to infected).
HPV transmits via sexual contact, and in HPVsim this is represented by sexual networks that allow agents to interact with one another. For the moment, HPVsim only models heterosexual partnerships. The sexual contact networks in HPVsim can have multiple contact layers, with each layer having different properties that characterize sexual contact, including the duration of the contact, age mixing preferences of partners, etc. HPVsim comes with two options for the sexual network:
The random option has a single contact layer. The number of partners that each agent has is Poisson distributed with a mean of 1.
The default option has 3 contact layers, representing marital, casual, and one-off partnership types.
Assortative mixing#
By default, new partnerships are formed based on age mixing patterns. There is an optional additional dimension of assortative mixing that could be used to represent geographical location or other demographic groups.
To enable the additional mixing, user needs to specify additional parameters that define 1) the number of clusters to use n_clusters; 2) a mixing matrix add_mixing that specifies pair-wise relative mixing weights between clusters.
[1]:
import sciris as sc
import numpy as np
import hpvsim as hpv
import scipy.linalg
hpv.options(jupyter=True, verbose=False)
base_pars = {
'n_agents': 2e4,
'start': 1970,
'end': 2020,
'location': 'nigeria',
'rand_seed': 1,
}
# Default: well-mixed (1 cluster)
sim0 = hpv.Sim(pars=base_pars)
assert sim0['n_clusters'] == 1
# Multiple clusters but well-mixed
pars1 = sc.dcp(base_pars)
pars1['n_clusters'] = 10 # create 10 clusters
pars1['add_mixing'] = np.ones((10, 10)) # set all between-cluster mixing to the same as within-cluster mixing
sim1 = hpv.Sim(pars=pars1)
print(sim1['add_mixing']) # print actual mixing matrix
# Modifying mixing matrix
pars2 = sc.dcp(base_pars)
pars2['n_clusters'] = 3 # create 3 clusters
pars2['add_mixing'] = np.array([[1, 0, 0],
[0, 1, 0],
[0, 0, 1]]) # No between-cluster mixing
sim2 = hpv.Sim(pars=pars2)
# Mixing by distance
pars3 = sc.dcp(base_pars)
pars3['n_clusters'] = 5
pars3['add_mixing'] = scipy.linalg.circulant([1,0.5,0.1,0.1,0.5])
sim3 = hpv.Sim(pars=pars3)
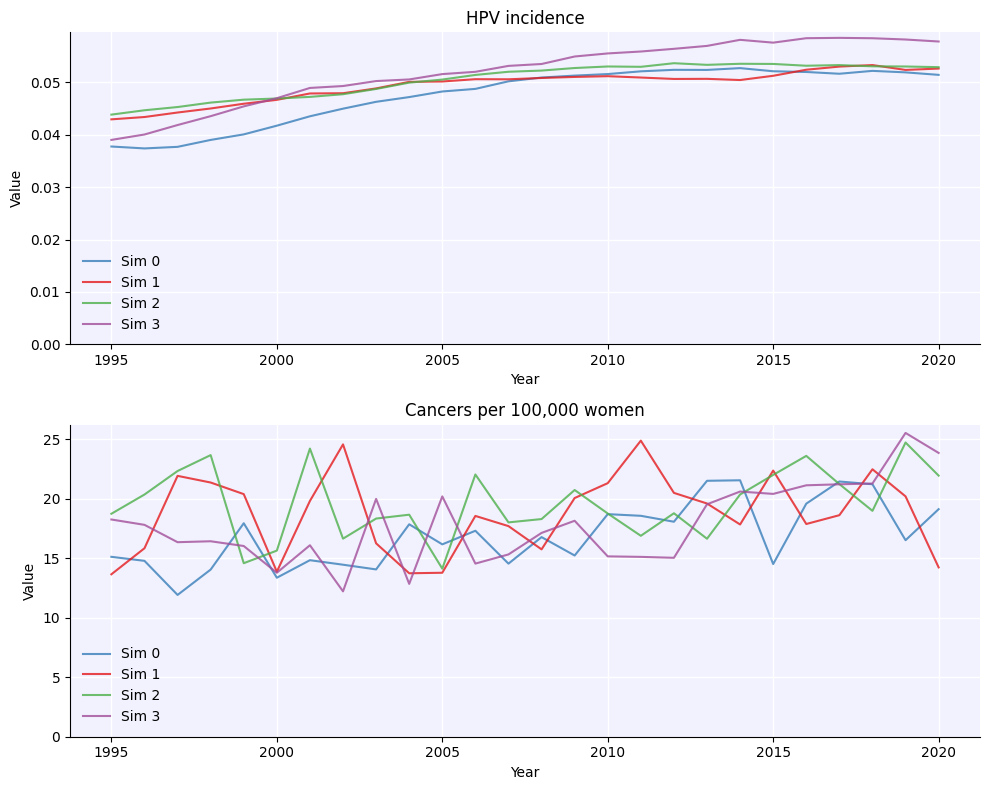
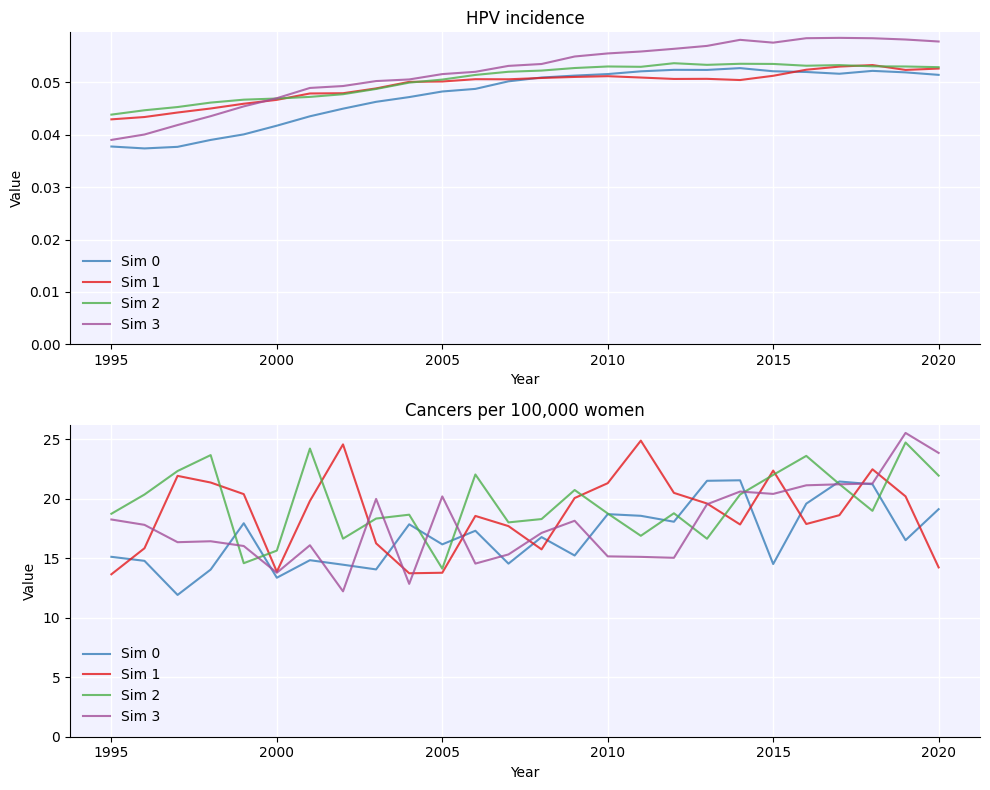
msim = hpv.parallel(sim0, sim1, sim2, sim3)
msim.plot()
HPVsim 2.1.0 (2025-03-25) — © 2023-2025 by IDM
Loading location-specific demographic data for "nigeria"
Loading location-specific demographic data for "nigeria"
[[1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]
[1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]
[1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]
[1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]
[1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]
[1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]
[1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]
[1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]
[1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]
[1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]]
Loading location-specific demographic data for "nigeria"
Loading location-specific demographic data for "nigeria"
[1]: