Model overview#
Disease models play an important role in understanding and managing the transmission dynamics of various pathogens. We can use them to describe the spatial and temporal patterns of disease prevalence, as well as to explore or better understand the factors that influence infection incidence. Modeling is a key step in understanding what treatments and interventions can be most effective, how cost-effective these approaches may be, and what specific factors need to be considered when trying to eradicate disease. The findings can be used to guide policy for implementing practical real-world solutions.
To understand the complex dynamics underlying disease transmission, epidemiologists often use a set of models called compartmental models. Developed in the early 20th century, these models stratify a population into groups, generally based on their risk or infection status. Underlying these models is a system of differential equations that track the number of people in each category over time. If you would like a more in-depth introduction to epidemiology and disease modeling, you may want to take the Epidemics course from The Pennsylvania State University through Coursera.
Agent-based models#
Epidemiological MODeling software (EMOD) is an agent-based model (ABM), another powerful tool used to help understand the complexity inherent in disease transmission systems. EMOD is a discrete time, Monte Carlo method simulator that simulates the simultaneous interactions of agents in an effort to recreate complex phenomena. Each agent (such as a human or vector) can be assigned a variety of properties (for example, age, gender, etc.), and their behavior and interactions with one another are determined using decision rules. EMOD and other ABMs must be run many times to produce probability distributions of potential outcomes, better capturing uncertainty than compartmental models.
These models have strong predictive power and are able to leverage spatial and temporal dynamics. Further, complex environments can be developed in which the agents act, and agents may “learn” from interactions or “adapt” to their environment. As a result, ABMs are excellent for identifying emerging properties of the system: patterns that are not explicitly modeled, but instead occur as a consequence of the rules that govern the agents.
EMOD can be calibrated to particular geographic locations, and the microsolver framework enables the model’s functionality to be highly modifiable. Further, the framework includes the ability to add intervention campaigns, and those interventions can be specified to target particular populations or sub-populations of human or vector groups. This intervention targeting is capable of simulating complex cascade of care systems. EMOD is very useful for determining the best intervention strategies to use in particular areas for burden reduction or elimination.
EMOD supports different simulation types for various diseases and transmission types. EMOD uses a layered architecture in which the base functionality is contained in the generic model and inherited by the transmission-level models that in turn are inherited by specific disease models. Because the EMOD modeling software can simulate all of these diseases, each “model” is more accurately referred to as a simulation type. For more information on the software architecture and inheritance, see Overview of EMOD software.
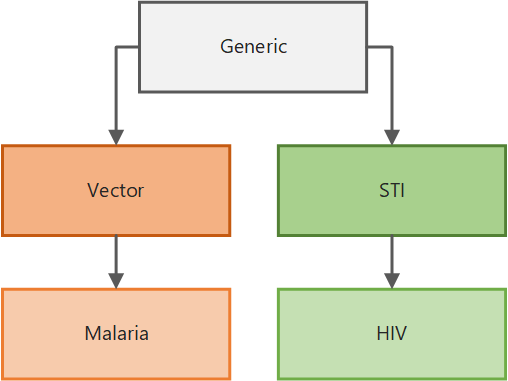
Simulation type inheritance#
The EMOD generic model forms the foundation for all other disease models in EMOD. It provides the fundamental logic for contact-based disease transmission and interventions aimed at controlling the spread of disease that are targeted to individuals or geographic nodes. You can easily add heterogeneity to your simulation by configuring he infectivity of the disease, susceptibility of individuals, and more.
The figure below demonstrates the main components of the generic EMOD simulation type. Individuals reside in geographic nodes and can migrate from one node to another. Infected individuals shed contagion into a pool that can infect susceptible individuals. When modeling malaria, vector and malaria parasite biology is added to the model to simulate disease transmission rather than a simple contact-based contagion pool. You can assign properties to individuals and nodes to vary how interventions are distributed.
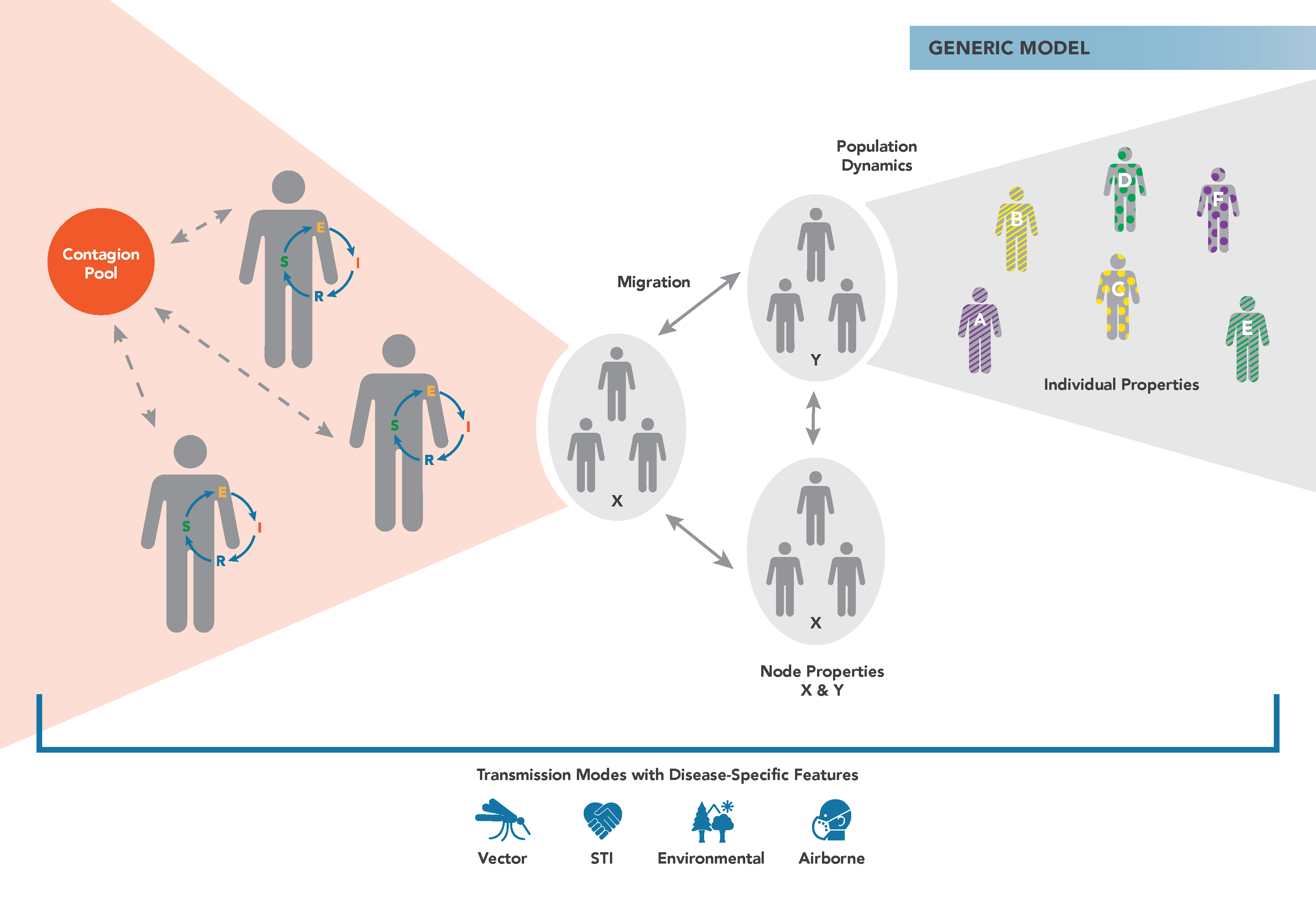
The configuration of the model regarding infectivity, immune response, and other qualities is handled via several JSON (JavaScript Object Notation) files. For more information, see Overview of EMOD software.
- Vector biology
- Malaria transmission and treatment
- Disease outbreaks, reservoirs, and endemicity
- Adding heterogeneity
- Creating campaigns
