SpatialReport#
The spatial output report breaks the channel data down per node, rather than across the entire simulation. It is a set of binary files, consisting of one file per channel. For each value set in the Spatial_Output_Channels configuration parameter array, a binary file with the name convention SpatialReport_<channel>.bin is generated. In addition, Enable_Spatial_Output must be set to 1.
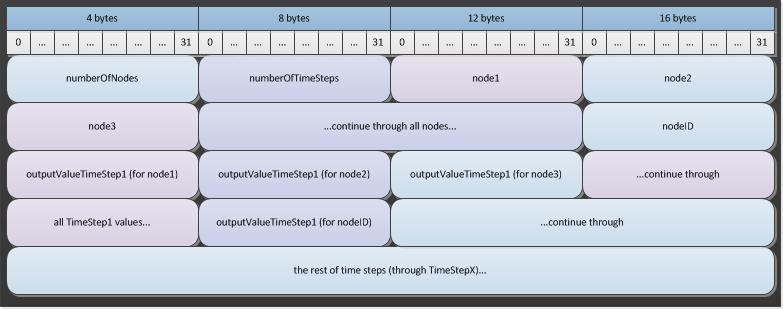
The binary format of the file consists of a stream of 4-byte integers followed by a stream of 4-byte floating point values. The first value is a 4-byte integer representing the number of nodes in the file and the second is a 4-byte integer that contains the number of time steps in the file. Following these two values is a stream of 4-byte integers that contain the node ID values in the order they will appear in the rest of the file. Following the node IDs is an array of 4-byte floating point values that represent the output values at the first time step for each node. The next array contains the values at the second time step, and so on.
The following diagram shows the format for data in the spatial output report file:
Configuration#
The following is an example of a spatial output channel configuration (config.json), and the following table defines the spatial output channels you can add to this report.
{
"Enable_Spatial_Output": 1,
"Spatial_Output_Channels": [
"Population",
"New_Infections",
"Prevalence",
"Births"
]
}
Output file data#
Channel |
Description |
|---|---|
Adult_Vectors |
The total number of adult female vectors in the node at each time step. Includes vectors from all species and all infectious sates (not infected, infected, and infectious). |
Air_Temperature |
The air temperature of that day for that node in Celcius. |
Births |
The live births cumulative up to that day for that node. |
Blood_Smear_Gametocyte_Prevalence |
The fraction of the population that is detectable with the BLOOD_SMEAR_GAMETOCYTES version of MalariaDiagnostic. The detectability of the diagnostic is controlled by the parameters Report_Gametocyte_Smear_Sensitivity and Report_Detection_Threshold_Blood_Smear_Gametocytes. |
Blood_Smear_Parasite_Prevalence |
The fraction of the population that is detectable with the BLOOD_SMEAR_PARASITES version of MalariaDiagnostic. The detectability of the diagnostic is controlled by the parameters Report_Parasite_Smear_Sensitivity and Report_Detection_Threshold_Blood_Smear_Parasites. |
Campaign_Cost |
The cumulative cost of the interventions that have been distributed to that node. This is based on the Cost_To_Consumer parameter in the interventions used. |
Daily_Bites_Per_Human |
The average number of mosquito bites received per individual on that day. |
Daily_EIR |
The number of infectious bites per person in the node made that day/reporting period as determined by the vector model. It includes the infectious bites of all species for both indoor and outdoor bites. |
Disease_Deaths |
The number of people that died in the node that day. The population for the node at the beginning of the next time step will be the population at this time step minus the number of deaths plus new births. |
Fever_Prevalence |
This fraction is the number of people whose fever is above the Report_Detection_Threshold_Fever threshold divided by the number of people in the node. |
Human_Infectious_Reservoir |
The sum of the infectiousness of each person in the node; a person’s infectiousness is the probability of infecting a mosquito during a successful blood meal. |
Infected |
The fraction of the population currently infected. |
Infectious Vectors |
The fraction of vectors in the simulation that are currently infected and infectious. |
Land_Temperature |
The temperature of the land/ground that day in that node in degrees Celsius. |
Mean_Parasitemia |
The geometric mean number of parasites per microliter of blood. |
New_Clinical_Cases |
The number of new clinical cases for a given day. |
New_Infections |
The number of people who received a new infection in that node on that time step. |
New_Reported_Infections |
The number of people reported to have received a new infection in that node on that time step. This number should be about 50% of the value in New_Infections. |
New_Severe_Cases |
The number of new severe cases of malaria on that day in the node. An individual is considered to be a severe case if the combined probability of anemia, parasite density, and fever is greater than a uniform random number. This combined probability is the combination of sigmoid using the following parameters: Anemia_Severe_Threshold and Anemia_Severe_Inverse_Width, Parasite_Severe_Threshold and Parasite_Severe_Inverse_Width, and Fever_Severe_Threshold and Fever_Severe_Inverse_Width. |
PCR_Gametocyte_Prevalence |
The fraction of the population that is detectable with the PCR_GAMETOCYTES version of MalariaDiagnostic. The detectability of the diagnostic is controlled by the parameter Report_Detection_Threshold_PCR_Gametocytes. |
PCR_Parasite_Prevalence |
The fraction of the population that is detectable with the PCR_PARASITES version of MalariaDiagnostic. The detectability of the diagnostic is controlled by the parameter Report_Detection_Threshold_PCR_Parasites. |
PfHRP2_Prevalence |
The fraction of the population that is detectable with the PF_HRP2 version of MalariaDiagnostic. The detectability of the diagnostic is controlled by the parameter Report_Detection_Threshold_PCR_PfHRP2. |
Population |
The total number of individuals in the node on that day. |
Prevalence |
The fraction of the population currently infected in that node on that day. |
Rainfall |
The number millimeters of rainfall in that node on that day. |
Relative_Humidity |
The amount of water vapor present in air expressed as a percentage of the amount needed for saturation at the same temperature for the node on that day. |
True_Prevalence |
The fraction of the population that is detectable with the TRUE_PARASITE_DENSITY version of MalariaDiagnostic. The detectability of the diagnostic is controlled by the parameter Report_Detection_Threshold_True_Parasite_Density. |
