Serialized population file#
To model a population with endemic disease, you cannot use a common modeling technique in which you introduce a disease outbreak to a naive population and analyze the immediate aftermath. Doing so would create a population missing the levels of natural immunity built up in the population due to the history of exposure to the pathogen. Instead, you must run the simulation for a period of time until the disease dynamics reach an equilibrium (aside from stochastic noise). This is known as steady state. The simulation output prior to that point is disregarded. This is a modeling concept known as simulation burn-in, borrowed from the electronics industry where the first items produced by a manufacturing process are discarded before the process is applied.
However, the time necessary to run simulations until this point can be significant, especially for large populations. Indeed, for endemic disease present at low absolute prevalence, you should simulate a larger population size that allows a small number of infected individuals to be represented.
EMOD avoids the need to run the burn-in period again and again with each simulation by using serialization to save the population state after it reaches equilibrium. Then, when you want to begin a subsequent simulation investigating the outcome of a particular set of interventions, you can begin the simulation at that point rather than needing to re-run the burn-in period. You can serialize the population at multiple time steps during a simulation.
The serialized population files created are placed in the output directory and use the naming convention state-<timestep>.dtk. They are binary files that contain state information about every agent in a simulation: their health status, age, property values, and more. These files can be consumed by subsequent simulations to decrease run time.
Note
If you used repeating interventions during the burn-in period, those interventions will not continue based on the information in the serialized population file. Check your campaign file for repeating interventions and reconfigure them as needed for the period after burn-in.
Not every type of intervention can be serialized. Please see Individual-level interventions and Node-level interventions for information on which interventions can be serialized.
See Simulation setup parameters for more information on configuration. See Serializing populations for more information on utilizing serialized populations in your simulations.
File format#
Serialized population files are saved in JSON format. The file structure reflects the hierarchical structure of the objects in Eradication.exe. Each file starts with a header and then contains information about the simulation and the nodes. The following figure gives a rough overview of the JSON file structure (note that only vector or malaria simulations will have larval habitats, vector populations, or vector life cycles).
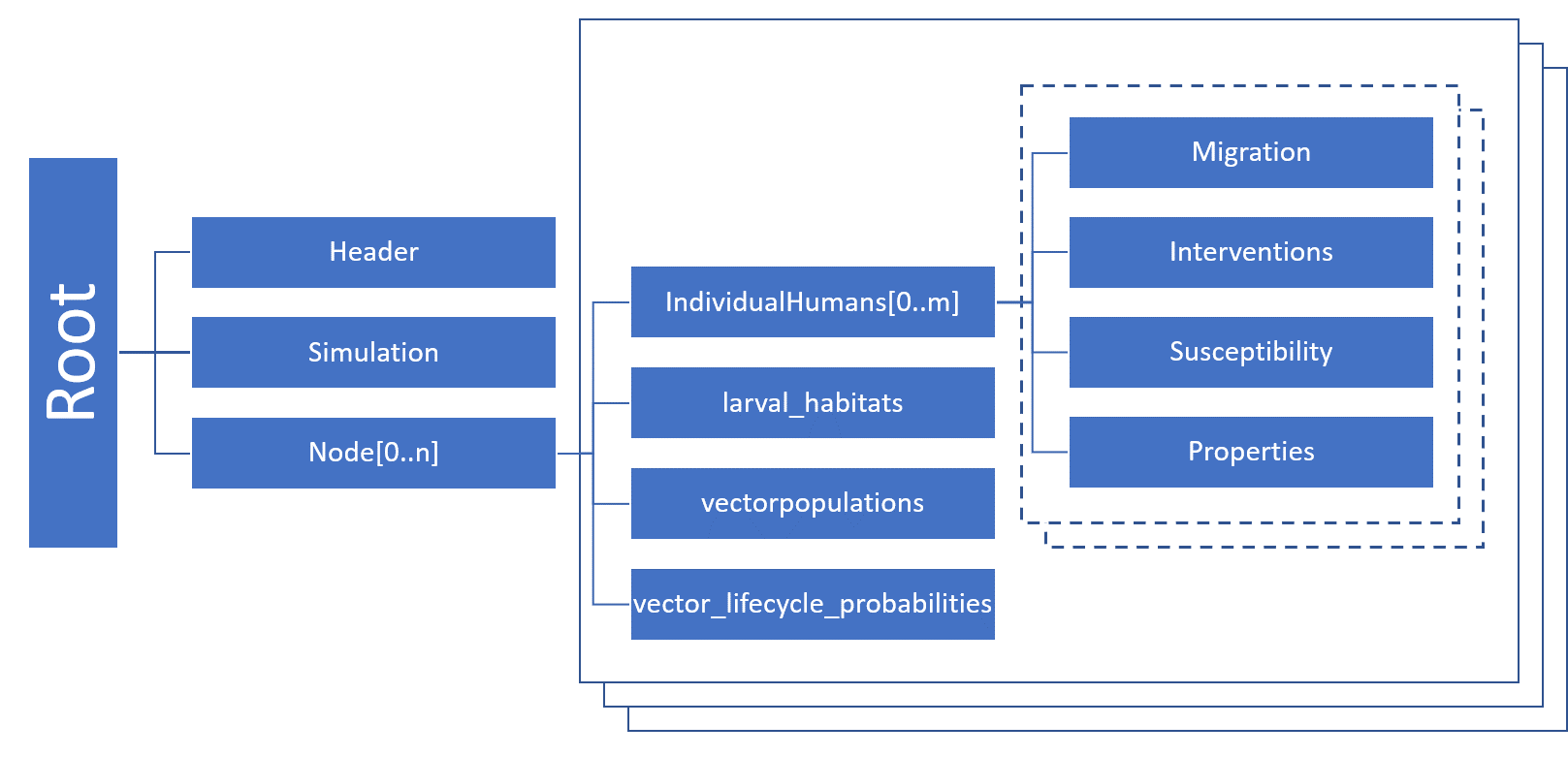
Internal state#
The structure of a serialized population file reflects the relations of objects in a simulation. On the highest level, a serialized population file contains a list with nodes, each node is the root of a tree. At the configured timestep the simulation calls Node::serialize() which then calls the serialized method of other members of this object. The Node class, for instance, contains a list with individuals, each individual has certain attributes and a list of infections. Each infection has certain attributes and might contain again a list of objects, etc. This continues until there are no more objects.
All objects that are instantiated from classes that inherit from ISerializable and implement the ISerializable interface can write itself to a buffer. It depends on the chosen writer how this buffer is written and where it is written. Here we assume that a JSON writer is used and that the writer adds variable names and formatting characters so that a valid JSON file is generated. A similar mechanism is at play when a simulation is loaded again from a file. The names of variables and their values are used to re-instantiate the same objects. Mind that EMOD is written in a compiled language, so the description of data and objects (classes and variable types) in the source code must match the data types in the serialized population file, e.g. if a variable is of type integer changing its value to a float won’t have the anticipated effect.
Multi-core#
If the simulation is run on several cores, each core saves one file at the configured timestep(s). To continue the simulation one EMOD file must be defined per node.
File Name#
The format for single core is: state-timestep.dtk e.g. state-00100.dtk
The format for multi-core is: state-timestep-core.dtk e.g. state-00100-001.dtk
Header#
Each EMOD file starts with a header in JSON format that contains information about the data in the file.
Version 2 and 3
Magic Number |
IDTK |
4 byte |
|
Header Size |
160 |
Integer |
Size of header in bytes |
Metadata
Parameter |
Example |
Type |
Description |
|---|---|---|---|
version |
2/3 |
Integer |
Version number |
date |
Day Mon day HH:MM:SS year |
String |
A string |
compressed |
True |
Boolean |
Content compressed |
engine |
LZ4 |
String |
Compression engine |
bytecount |
285140 |
Integer |
Total number of bytes in file |
chunkcount |
3 |
Integer |
Number of chunks |
chunksizes |
[368,141552,143220] |
List of Integers |
List of chunk sizes |
Version 4
Magic Number |
IDTK |
4 byte |
|
Header Size |
160 |
Integer |
Size of header in bytes |
Metadata
Parameter |
Example |
Type |
Description |
|---|---|---|---|
version |
4 |
Integer |
Version number |
date |
Day Mon day HH:MM:SS year |
String |
A string |
compression |
LZ4 |
NONE, LZ4, SNAPPY |
Content compressed |
bytecount |
285140 |
Integer |
Total number of bytes in file |
chunkcount |
3 |
Integer |
Number of chunks |
chunksizes |
[368,141552,143220] |
List of Integers |
List of chunk sizes |
